β-Catenin/c-Myc Axis Modulates Autophagy Response to Different Ammonia Concentrations
- PMID: 39798123
- PMCID: PMC11911958
- DOI: 10.1002/adbi.202400408
β-Catenin/c-Myc Axis Modulates Autophagy Response to Different Ammonia Concentrations
Abstract
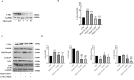
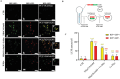
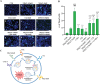
Ammonia a by-product of nitrogen containing molecules is detoxified by liver into non-toxic urea and glutamine. Impaired ammonia detoxification leads to hyperammonemia. Ammonia has a dual role on autophagy, it acts as inducer at low concentrations and as inhibitor at high concentrations. However, little is known about the mechanisms responsible for this switch. Wnt/β-catenin signalling is emerging for its role in the regulation of ammonia metabolizing enzymes and autophagosome synthesis through c-Myc. Here, using Huh7 cell line, we show a modulation in c-Myc expression under different ammonia concentrations. An increase in c-Myc expression and in its transcriptional regulator β-catenin was detected at low concentrations of ammonia, when autophagy is active, whereas these modifications were lost under high ammonia concentrations. These observations were also recapitulated in the livers of spf-ash mice, a model of constitutive hyperammonaemia due to deficiency in ornithine transcarbamylase enzyme. Moreover, c-Myc-mediated activation of autophagy plays a cytoprotective role in cells under ammonia stress conditions as confirmed through the pharmacological inhibition of c-Myc in Huh7 cells treated with low ammonia concentrations. In conclusion, the unravelled role of c-Myc in modulating ammonia induced autophagy opens new landscapes for the development of novel strategies for the treatment of hyperammonemia.
Keywords: autophagy; c‐Myc; hyperammonemia; liver.
© 2025 The Author(s). Advanced Biology published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Eng C. H., Yu K., Lucas J., White E., Abraham R. T., Sci. Signaling 2010, 3, ra31. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

