The steroid hormone 20-hydroxyecdysone induces lipophagy via the brain-adipose tissue axis by promoting the adipokinetic hormone pathway
- PMID: 39798879
- PMCID: PMC11835591
- DOI: 10.1016/j.jbc.2025.108179
The steroid hormone 20-hydroxyecdysone induces lipophagy via the brain-adipose tissue axis by promoting the adipokinetic hormone pathway
Abstract
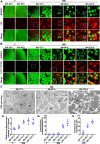
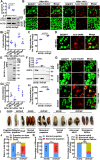
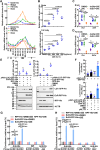
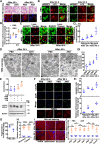
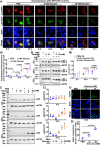
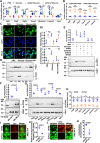
Lipophagy is a way to degrade lipids; however, the molecular mechanisms are not fully understood. Using the holometabolous lepidopteran insect Helicoverpa armigera, cotton bollworm, as a model, we revealed that the larval fat body undergoes lipophagy during metamorphosis, and lipophagy is essential for metamorphosis. The steroid hormone 20-hydroxyecdysone (20E) induced lipophagy by promoting the expression of the peptide hormone adipokinetic hormone (AKH, the insect analog of glucagon) and the adipokinetic hormone receptor (AKHR). Akh was highly expressed in the brain and Akhr was expressed in various tissues. The 20E upregulated the expression of Akh and Akhr by its nuclear receptor EcR during metamorphosis. AKH and AKHR increased glucose levels via gluconeogenesis and promoted lipophagy. The high glucose level induced acetylation of FOXO and nuclear localization to promote the expression of lipases and autophagy genes. Thus, the steroid hormone 20E induced lipophagy via the brain-adipose tissue axis by promoting the AKH pathway, which presented nutrients and energy to pupal and adult development during insect metamorphosis after feeding stops.
Keywords: 20-hydroxyecdysone; adipokinetic hormone; adipokinetic hormone receptor; glucose; lipophagy.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Raju B., Cryer P.E. Maintenance of the postabsorptive plasma glucose concentration: insulin or insulin plus glucagon? Am. J. Physiol. Endocrinol. Metab. 2005;289:E181–E186. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

