Promoted read-through and mutation against pseudouridine-CMC by an evolved reverse transcriptase
- PMID: 39799263
- PMCID: PMC11724883
- DOI: 10.1038/s42003-025-07467-4
Promoted read-through and mutation against pseudouridine-CMC by an evolved reverse transcriptase
Abstract
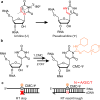
Pseudouridine (Ψ) is an abundant RNA chemical modification that plays critical biological functions. Current Ψ detection methods are limited in identifying Ψs at base-resolution in U-rich sequence contexts, where Ψ occurs frequently. Here we report "Mut-Ψ-seq" that utilizes the classic N-cyclohexyl N'-(2-morpholinoethyl)carbodiimide (CMC) agent and an evolved reverse transcriptase ("RT-1306") for Ψ mapping at base-resolution. CMC selectively labels Ψs in RNA forming the CMC-Ψ adduct and we show that RT-1306 presents promoted read-through and mutation against the CMC-Ψ. We report a high-confidence list of Ψ sites in polyA-enriched RNAs from HEK-293T cells identified by orthogonal chemical treatments (CMC and bisulfite). The mutation signatures resolve the position of Ψ in UU-containing sequences, revealing diverse occurrence of Ψs in such sequences. This work provides methods and datasets for biological research of Ψ, and expands the toolkit for epitranscriptomic studies by combining the reverse transcriptase engineering and selective chemical labeling strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Update of
-
Promoted Read-through and Mutation Against Pseudouridine-CMC by an Evolved Reverse Transcriptase.bioRxiv [Preprint]. 2024 Jul 3:2024.07.03.601893. doi: 10.1101/2024.07.03.601893. bioRxiv. 2024. Update in: Commun Biol. 2025 Jan 11;8(1):40. doi: 10.1038/s42003-025-07467-4. PMID: 39005393 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

