PhyloMix: enhancing microbiome-trait association prediction through phylogeny-mixing augmentation
- PMID: 39799515
- PMCID: PMC11849959
- DOI: 10.1093/bioinformatics/btaf014
PhyloMix: enhancing microbiome-trait association prediction through phylogeny-mixing augmentation
Abstract
Motivation: Understanding the associations between traits and microbial composition is a fundamental objective in microbiome research. Recently, researchers have turned to machine learning (ML) models to achieve this goal with promising results. However, the effectiveness of advanced ML models is often limited by the unique characteristics of microbiome data, which are typically high-dimensional, compositional, and imbalanced. These characteristics can hinder the models' ability to fully explore the relationships among taxa in predictive analyses. To address this challenge, data augmentation has become crucial. It involves generating synthetic samples with artificial labels based on existing data and incorporating these samples into the training set to improve ML model performance.
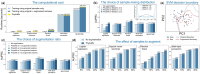
Results: Here, we propose PhyloMix, a novel data augmentation method specifically designed for microbiome data to enhance predictive analyses. PhyloMix leverages the phylogenetic relationships among microbiome taxa as an informative prior to guide the generation of synthetic microbial samples. Leveraging phylogeny, PhyloMix creates new samples by removing a subtree from one sample and combining it with the corresponding subtree from another sample. Notably, PhyloMix is designed to address the compositional nature of microbiome data, effectively handling both raw counts and relative abundances. This approach introduces sufficient diversity into the augmented samples, leading to improved predictive performance. We empirically evaluated PhyloMix on six real microbiome datasets across five commonly used ML models. PhyloMix significantly outperforms distinct baseline methods including sample-mixing-based data augmentation techniques like vanilla mixup and compositional cutmix, as well as the phylogeny-based method TADA. We also demonstrated the wide applicability of PhyloMix in both supervised learning and contrastive representation learning.
Availability and implementation: The Apache-licensed source code is available at (https://github.com/batmen-lab/phylomix).
© The Author(s) 2025. Published by Oxford University Press.
Figures


Similar articles
-
TADA: phylogenetic augmentation of microbiome samples enhances phenotype classification.Bioinformatics. 2019 Jul 15;35(14):i31-i40. doi: 10.1093/bioinformatics/btz394. Bioinformatics. 2019. PMID: 31510701 Free PMC article.
-
PolypMixNet: Enhancing semi-supervised polyp segmentation with polyp-aware augmentation.Comput Biol Med. 2024 Mar;170:108006. doi: 10.1016/j.compbiomed.2024.108006. Epub 2024 Jan 15. Comput Biol Med. 2024. PMID: 38325216
-
Transformation and differential abundance analysis of microbiome data incorporating phylogeny.Bioinformatics. 2021 Dec 11;37(24):4652-4660. doi: 10.1093/bioinformatics/btab543. Bioinformatics. 2021. PMID: 34302462
-
Techniques for learning and transferring knowledge for microbiome-based classification and prediction: review and assessment.Brief Bioinform. 2024 Nov 22;26(1):bbaf015. doi: 10.1093/bib/bbaf015. Brief Bioinform. 2024. PMID: 39820436 Free PMC article. Review.
-
Methodology for microbiome data analysis: An overview.Comput Biol Med. 2025 Jun;192(Pt A):110157. doi: 10.1016/j.compbiomed.2025.110157. Epub 2025 Apr 24. Comput Biol Med. 2025. PMID: 40279974 Review.
References
-
- Adebayo J, Gilmer J, Muelly M et al. Sanity checks for saliency maps. In: Proceedings of the 32nd International Conference on Neural Information Processing Systems, Montréal Canada, 2013, pp. 9525–36. Red Hook, NY, United States: Curran Associates Inc., 2018.
-
- Aitchison J. The statistical analysis of compositional data. J R Stat Soc Ser B (Methodol) 1982;44:139–60.
-
- Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 2013;35:1798–828. - PubMed
-
- Boktor JC, Sharon G, Verhagen Metman LA et al. Integrated multi-cohort analysis of the Parkinson’s disease gut metagenome. Mov Disord 2023;38:399–409. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

