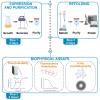
Protocol for high-yield bacterial expression and purification of the voltage-dependent anion channel 1 for high-throughput biophysical assays
- PMID: 39799574
- PMCID: PMC11772971
- DOI: 10.1016/j.xpro.2024.103557
Protocol for high-yield bacterial expression and purification of the voltage-dependent anion channel 1 for high-throughput biophysical assays
Abstract
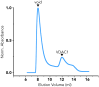
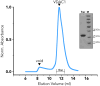
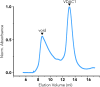
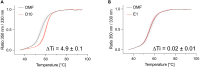
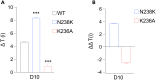
Voltage-dependent anion channel 1 (VDAC1) is a key protein in cellular metabolism and apoptosis. Here, we present a protocol to express and purify milligram amounts of recombinant VDAC1 in Escherichia coli. We detail steps for a fluorescence polarization-based high-throughput screening assay using NADH displacement, along with procedures for thermostability, fluorescence polarization, and X-ray crystallography. In this context, we demonstrate how 2-methyl-2,4-pentanediol (MPD), a crystallization reagent, interferes with VDAC1 small-molecule binding, hindering the detection of these ligands in the crystal. For complete details on the use and execution of this protocol, please refer to Conti Nibali et al.1.
Keywords: Biophysics; Protein Biochemistry; Protein expression and purification; X-ray Crystallography.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Conti Nibali S., De Siervi S., Luchinat E., Magrì A., Messina A., Brocca L., Mantovani S., Oliviero B., Mondelli M.U., De Pinto V., et al. VDAC1-interacting molecules promote cell death in cancer organoids through mitochondrial-dependent metabolic interference. iScience. 2024;27 doi: 10.1016/j.isci.2024.109853. - DOI - PMC - PubMed
-
- Ujwal R., Cascio D., Colletier J.P., Faham S., Zhang J., Toro L., Ping P., Abramson J. The crystal structure of mouse VDAC1 at 2.3 (2008). A resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. USA. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

