Radiomics signature for dynamic monitoring of tumor inflamed microenvironment and immunotherapy response prediction
- PMID: 39800381
- PMCID: PMC11749429
- DOI: 10.1136/jitc-2024-009140
Radiomics signature for dynamic monitoring of tumor inflamed microenvironment and immunotherapy response prediction
Abstract
Background: The efficacy of immune checkpoint inhibitors (ICIs) depends on the tumor immune microenvironment (TIME), with a preference for a T cell-inflamed TIME. However, challenges in tissue-based assessments via biopsies have triggered the exploration of non-invasive alternatives, such as radiomics, to comprehensively evaluate TIME across diverse cancers. To address these challenges, we develop an ICI response signature by integrating radiomics with T cell-inflamed gene-expression profiles.
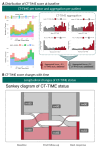
Methods: We conducted a pan-cancer investigation into the utility of radiomics for TIME assessment, including 1360 tumors from 428 patients. Leveraging contrast-enhanced CT images, we characterized TIME through RNA gene expression analysis, using the T cell-inflamed gene expression signature. Subsequently, a pan-cancer CT-radiomic signature predicting inflamed TIME (CT-TIME) was developed and externally validated. Machine learning was employed to select robust radiomic features and predict inflamed TIME. The study also integrated independent cohorts with longitudinal CT images, baseline biopsies, and comprehensive immunohistochemistry panel evaluation to assess the pan-cancer biological associations, spatiotemporal landscape and clinical utility of the CT-TIME.
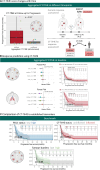
Results: The CT-TIME signature, comprising four radiomic features linked to a T-cell inflamed microenvironment, demonstrated robust performance with AUCs (95% CI) of 0.85 (0.73 to 0.96) (training) and 0.78 (0.65 to 0.92) (external validation). CT-TIME scores exhibited positive correlations with CD3, CD8, and CD163 expression. Intrapatient analysis revealed considerable heterogeneity in TIME between tumors, which could not be assessed using biopsies. Evaluation of aggregated per-patient CT-TIME scores highlighted its promising clinical utility for dynamically assessing the immune microenvironment and predicting immunotherapy response across diverse scenarios in advanced cancer. Despite demonstrating progression disease at the first follow-up, patients within the inflamed status group, identified by CT-TIME, exhibited significantly prolonged progression-free survival (PFS), with some surpassing 5 months, suggesting a potential phenomenon of pseudoprogression. Cox models using aggregated CT-TIME scores from baseline images revealed a statistically significant reduction in the risk of PFS in the pan-cancer cohort (HR 0.62, 95% CI 0.44 to 0.88, p=0.007), and Kaplan-Meier analysis further confirmed substantial differences in PFS between patients with inflamed and uninflamed status (log-rank test p=0.009).
Conclusions: The signature holds promise for impacting clinical decision-making, pan-cancer patient stratification, and treatment outcomes in immune checkpoint therapies.
Keywords: Biomarker; Computed tomography; Immunotherapy; Pseudoprogression; Tumor microenvironment - TME.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: KB, RA, ME, JF, FG, ML, OP, GS, and CZ—nothing to disclose. EF—consulting fees: Abbvie, Amgen, Astra Zeneca, Bayer, Beigene, Boehringer Ingelheim, Bristol Myers Squibb, Eli Lilly, F. Hoffmann-La Roche, Gilead, Glaxo Smith Kline, Janssen, Merck Serono, Merck Sharp & Dohme, Novartis, Peptomyc, Pfizer, Regeneron, Sanofi, Takeda, Turning Point, Daiichi Sankyo. Payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events: Amgen, Astra Zeneca, Bristol Myers Squibb, Daiichi Sankyo, Eli Lilly, F. Hoffmann – La Roche, Genentech, Janssen, Medical Trends, Medscape, Merck Serono, Merck Sharp & Dohme, Peervoice, Pfizer, Sanofi, Takeda, Touch Oncology. Support for attending meetings and/or travel: Astra Zeneca, Janssen, Roche. EG: grants or contracts from any entity: Novartis, Roche, Thermo Fisher, AstraZeneca, Taiho, BeiGene, Janssen. Consulting fees: Roche, Ellipses Pharma, Boehringer Ingelheim, Janssen Global Services, Seattle Genetics, Thermo Fisher, MabDiscovery, Anaveon, F-Star Therapeutics, Hengrui, Sanofi, Incyte, Medscape. Payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events: Merck Sharp & Dohme, Roche, Thermo Fisher, Novartis, SeaGen. Stock or stock options: 1TRIALSP. PI or Co-PI—Institutional: Adaptimmune—Affimed—Amgen SA—Anaveon AG—AstraZeneca AB—Bicycletx—BioInvent International AB—Biontech SE—Biontech Small Molecules—Boehringer Ingelhem International—Catalym—Cyclacel Biopharmaceuticals—Cytovation AS—Cytomx—F.Hoffmann La Roche—F-Star Beta—Genentech—Genmab B.V—Hifibio Therapeutics—Hutchison Medipharma —Icon—Imcheck Therapeutics—Immunocore—Incyte Corporation—Incyte Europe Sàrl—Janssen-Cilag International NV—Janssen-Cilag SA—Laboratorios Servier SL—Medimmune—Merck & Co—Merck Kgga—Novartis Farmacéutica, S.A—Peptomyc—Pfizer Slu—Relay Therapeutics—Replimmune - Ribon Therapeutics—Ryvu Therapeutics SA—Seattle Genetics—Sotio as—Sqz Biotechnologies—Symphogen A/S—Taiho Pharma Usa—T-Knife Gmbh, NEXT Oncology. PN: payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events: NOVARTIS, Invited Speaker. Participation on a Data Safety Monitoring Board or Advisory Board: Bayer, Advisory Board, MSD ONCOLOGY, Advisory Board. Other financial or non-financial interests: Targos Molecular Pathology GmbH, Other, Consultant. RP-L: all support for the present manuscript: Astra Zeneca PoC Grant, CRIS Cancer Foundation. Consulting fees: Roche Pharma, Steering Committee Member of the ATHECA clinical trial. Leadership or fiduciary role in another board, society, committee or advocacy group, paid or unpaid: Cancer Core Europe Imaging Task Force Leader. RT: grants or contracts from any entity: Research grants from Astrazeneca, Beigene.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
