This is a preprint.
Demystifying Large Language Models for Medicine: A Primer
- PMID: 39801619
- PMCID: PMC11722506
Demystifying Large Language Models for Medicine: A Primer
Abstract
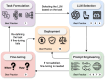
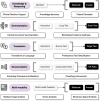
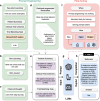
Large language models (LLMs) represent a transformative class of AI tools capable of revolutionizing various aspects of healthcare by generating human-like responses across diverse contexts and adapting to novel tasks following human instructions. Their potential application spans a broad range of medical tasks, such as clinical documentation, matching patients to clinical trials, and answering medical questions. In this primer paper, we propose an actionable guideline to help healthcare professionals more efficiently utilize LLMs in their work, along with a set of best practices. This approach consists of several main phases, including formulating the task, choosing LLMs, prompt engineering, fine-tuning, and deployment. We start with the discussion of critical considerations in identifying healthcare tasks that align with the core capabilities of LLMs and selecting models based on the selected task and data, performance requirements, and model interface. We then review the strategies, such as prompt engineering and fine-tuning, to adapt standard LLMs to specialized medical tasks. Deployment considerations, including regulatory compliance, ethical guidelines, and continuous monitoring for fairness and bias, are also discussed. By providing a structured step-by-step methodology, this tutorial aims to equip healthcare professionals with the tools necessary to effectively integrate LLMs into clinical practice, ensuring that these powerful technologies are applied in a safe, reliable, and impactful manner.
Conflict of interest statement
Disclosures The recommendations in this article are those of the authors and do not necessarily represent the official position of the National Institutes of Health.
Figures

Similar articles
-
Utilizing large language models for gastroenterology research: a conceptual framework.Therap Adv Gastroenterol. 2025 Apr 1;18:17562848251328577. doi: 10.1177/17562848251328577. eCollection 2025. Therap Adv Gastroenterol. 2025. PMID: 40171241 Free PMC article. Review.
-
A Review of Large Language Models in Medical Education, Clinical Decision Support, and Healthcare Administration.Healthcare (Basel). 2025 Mar 10;13(6):603. doi: 10.3390/healthcare13060603. Healthcare (Basel). 2025. PMID: 40150453 Free PMC article. Review.
-
Leveraging Large Language Models for Precision Monitoring of Chemotherapy-Induced Toxicities: A Pilot Study with Expert Comparisons and Future Directions.Cancers (Basel). 2024 Aug 12;16(16):2830. doi: 10.3390/cancers16162830. Cancers (Basel). 2024. PMID: 39199603 Free PMC article.
-
Bridging the gap: a practical step-by-step approach to warrant safe implementation of large language models in healthcare.Front Artif Intell. 2025 Jan 27;8:1504805. doi: 10.3389/frai.2025.1504805. eCollection 2025. Front Artif Intell. 2025. PMID: 39931218 Free PMC article.
-
Prompt Engineering as an Important Emerging Skill for Medical Professionals: Tutorial.J Med Internet Res. 2023 Oct 4;25:e50638. doi: 10.2196/50638. J Med Internet Res. 2023. PMID: 37792434 Free PMC article.
References
-
- Achiam J., et al. Gpt-4 technical report. arXiv preprint arXiv:2303.08774 (2023).
-
- Anthropic A. The claude 3 model family: Opus, sonnet, haiku. Claude-3 Model Card (2024).
-
- Reid M. et al. Gemini 1.5: Unlocking multimodal understanding across millions of tokens of context. arXiv preprint arXiv:2403.05530 (2024).
-
- The llama 3 herd of models. arXiv preprint arXiv:2407.21783 (2024.
-
- Brown T., et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 33, 1877–1901 (2020).
Publication types
LinkOut - more resources
Full Text Sources
