QOMIC: quantum optimization for motif identification
- PMID: 39801778
- PMCID: PMC11725347
- DOI: 10.1093/bioadv/vbae208
QOMIC: quantum optimization for motif identification
Abstract
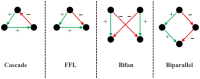
Motivation: Network motif identification (MI) problem aims to find topological patterns in biological networks. Identifying disjoint motifs is a computationally challenging problem using classical computers. Quantum computers enable solving high complexity problems which do not scale using classical computers. In this article, we develop the first quantum solution, called QOMIC (Quantum Optimization for Motif IdentifiCation), to the MI problem. QOMIC transforms the MI problem using a integer model, which serves as the foundation to develop our quantum solution. We develop and implement the quantum circuit to find motif locations in the given network using this model.
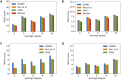
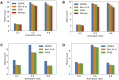
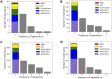
Results: Our experiments demonstrate that QOMIC outperforms the existing solutions developed for the classical computer, in term of motif counts. We also observe that QOMIC can efficiently find motifs in human regulatory networks associated with five neurodegenerative diseases: Alzheimer's, Parkinson's, Huntington's, Amyotrophic Lateral Sclerosis, and Motor Neurone Disease.
Availability and implementation: Our implementation can be found in https://github.com/ngominhhoang/Quantum-Motif-Identification.git.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures

References
-
- Aaronson S, Arkhipov A. The computational complexity of linear optics. In: Proceedings of the Forty-Third Annual ACM Symposium on Theory of Cmputing (STOC’11). New York, NY, USA: Association for Computing Machinery, 2011, 333–42.
-
- Aleksandrowicz G, Alexander T, Kl P et al. Qiskit: An Open-source Framework for Quantum Computing. 2019.
-
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet 2007;8:450–61. - PubMed
-
- Cerezo M, Arrasmith A, Babbush R et al. Variational quantum algorithms. Nat Rev Phys 2021;3:625–44.
LinkOut - more resources
Full Text Sources

