Deep learning segmentation architectures for automatic detection of pancreatic ductal adenocarcinoma in EUS-guided fine-needle biopsy samples based on whole-slide imaging
- PMID: 39802107
- PMCID: PMC11723688
- DOI: 10.1097/eus.0000000000000094
Deep learning segmentation architectures for automatic detection of pancreatic ductal adenocarcinoma in EUS-guided fine-needle biopsy samples based on whole-slide imaging
Abstract
Background: EUS-guided fine-needle biopsy is the procedure of choice for the diagnosis of pancreatic ductal adenocarcinoma (PDAC). Nevertheless, the samples obtained are small and require expertise in pathology, whereas the diagnosis is difficult in view of the scarcity of malignant cells and the important desmoplastic reaction of these tumors. With the help of artificial intelligence, the deep learning architectures produce a fast, accurate, and automated approach for PDAC image segmentation based on whole-slide imaging. Given the effectiveness of U-Net in semantic segmentation, numerous variants and improvements have emerged, specifically for whole-slide imaging segmentation.
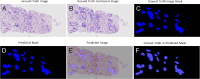
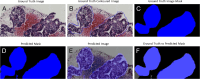
Methods: In this study, a comparison of 7 U-Net architecture variants was performed on 2 different datasets of EUS-guided fine-needle biopsy samples from 2 medical centers (31 and 33 whole-slide images, respectively) with different parameters and acquisition tools. The U-Net architecture variants evaluated included some that had not been previously explored for PDAC whole-slide image segmentation. The evaluation of their performance involved calculating accuracy through the mean Dice coefficient and mean intersection over union (IoU).
Results: The highest segmentation accuracies were obtained using Inception U-Net architecture for both datasets. PDAC tissue was segmented with the overall average Dice coefficient of 97.82% and IoU of 0.87 for Dataset 1, respectively, overall average Dice coefficient of 95.70%, and IoU of 0.79 for Dataset 2. Also, we considered the external testing of the trained segmentation models by performing the cross evaluations between the 2 datasets. The Inception U-Net model trained on Train Dataset 1 performed with the overall average Dice coefficient of 93.12% and IoU of 0.74 on Test Dataset 2. The Inception U-Net model trained on Train Dataset 2 performed with the overall average Dice coefficient of 92.09% and IoU of 0.81 on Test Dataset 1.
Conclusions: The findings of this study demonstrated the feasibility of utilizing artificial intelligence for assessing PDAC segmentation in whole-slide imaging, supported by promising scores.
Keywords: Artificial intelligence; Deep learning; EUS-guided fine-needle biopsy; Pancreatic ductal adenocarcinoma; Whole-slide imaging.
Copyright © 2024 The Author(s). Published by Wolters Kluwer Health, Inc on behalf of Scholar Media Publishing.
Conflict of interest statement
Adrian Săftoiu is an Associate Editor of the journal. The article was subjected to the standard procedures of the journal, with a review process independent of the editor and his research group.
Figures






References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin 2023;73(1):17–48. - PubMed
LinkOut - more resources
Full Text Sources

