The molecular features of non-peptidic nucleophilic substrates and acceptor proteins determine the efficiency of sortagging
- PMID: 39802631
- PMCID: PMC11721432
- DOI: 10.1039/d4cb00246f
The molecular features of non-peptidic nucleophilic substrates and acceptor proteins determine the efficiency of sortagging
Abstract
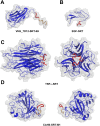
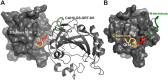
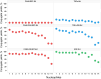
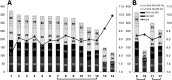
Sortase A-mediated ligation (SML) or "sortagging" has become a popular technology to selectively introduce structurally diverse protein modifications. Despite the great progress in the optimization of the reaction conditions and design of miscellaneous C- or N-terminal protein modification strategies, the reported yields of conjugates are highly variable. In this study, we have systematically investigated C-terminal protein sortagging efficiency using a combination of several rationally selected and modified acceptor proteins and a panel of incoming surrogate non-peptidic amine nucleophile substrates varying in the structural features of their amino linker parts and cargo molecules. Our data suggest that the sortagging efficiency is modulated by the combination of molecular features of the incoming nucleophilic substrate, including the ionization properties of the reactive amino group, structural recognition of the nucleophilic amino linker by the enzyme, as well as the molecular nature of the attached payload moiety. Previous reports have confirmed that the steric accessibility of the C-terminal SrtA recognition site in the acceptor protein is also the critical determinant of sortase reaction efficiency. We suggest a computational procedure for simplifying a priori predictions of sortagging outcomes through the structural assessment of the acceptor protein and introduction of a peptide linker, if deemed necessary.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures

References
LinkOut - more resources
Full Text Sources

