This is a preprint.
Transcriptome-wide outlier approach identifies individuals with minor spliceopathies
- PMID: 39802771
- PMCID: PMC11722475
- DOI: 10.1101/2025.01.02.24318941
Transcriptome-wide outlier approach identifies individuals with minor spliceopathies
Update in
-
Transcriptome-wide outlier approach identifies individuals with minor spliceopathies.Am J Hum Genet. 2025 Oct 2;112(10):2458-2475. doi: 10.1016/j.ajhg.2025.08.018. Epub 2025 Sep 19. Am J Hum Genet. 2025. PMID: 40975062
Abstract
RNA-sequencing has improved the diagnostic yield of individuals with rare diseases. Current analyses predominantly focus on identifying outliers in single genes that can be attributed to cis-acting variants within the gene locus. This approach overlooks causal variants with trans-acting effects on splicing transcriptome-wide, such as variants impacting spliceosome function. We present a transcriptomics-first method to diagnose individuals with rare diseases by examining transcriptome-wide patterns of splicing outliers. Using splicing outlier detection methods (FRASER and FRASER2) we characterized splicing outliers from whole blood for 390 individuals from the Genomics Research to Elucidate the Genetics of Rare Diseases (GREGoR) and Undiagnosed Diseases Network (UDN) consortia. We examined all samples for excess intron retention outliers in minor intron containing genes (MIGs). Minor introns, which make up about 0.5% of all introns in the human genome, are removed by small nuclear RNAs (snRNAs) in the minor spliceosome. This approach identified five individuals with excess intron retention outliers in MIGs, all of which were found to harbor rare, biallelic variants in minor spliceosome snRNAs. Four individuals had rare, compound heterozygous variants in RNU4ATAC, which aided the reclassification of four variants. Additionally, one individual had rare, highly conserved, compound heterozygous variants in RNU6ATAC that may disrupt the formation of the catalytic spliceosome, suggesting a novel gene-disease candidate. These results demonstrate that examining RNA-sequencing data for transcriptome-wide signatures can increase the diagnostic yield of individuals with rare diseases, provide variant-to-function interpretation of spliceopathies, and uncover novel disease gene associations.
Conflict of interest statement
DECLARATION OF INTERESTS AODL was a paid consultant for Tome Biosciences, Ono Pharma USA, and Addition Therapeutics. SBM is an advisor to Character Bio, Myome, PhiTech and Tenaya Therapeutics.
Figures

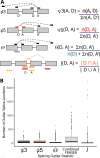

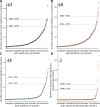


References
-
- Health T. L. G. The landscape for rare diseases in 2024. Lancet Glob. Health 12, e341 (2024). - PubMed
-
- European Union. REGULATION (EC) No 141/2000 OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL of 16 December 1999 on orphan medicinal products. Off. J. Eur. Communities (2000).
-
- Hartley T. et al. New Diagnostic Approaches for Undiagnosed Rare Genetic Diseases. Annu. Rev. Genomics Hum. Genet. 21, 351–372 (2020). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
