Efficient Hydrolysis of Fish Parvalbumin by Marine Bacterial Protease VSP2V-280: Allergen Removal
- PMID: 39803265
- PMCID: PMC11717067
- DOI: 10.1002/fsn3.4729
Efficient Hydrolysis of Fish Parvalbumin by Marine Bacterial Protease VSP2V-280: Allergen Removal
Abstract
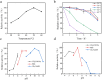
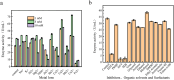
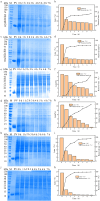
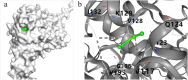
Parvalbumin is a major allergen in fish. However, there is currently no effective and safe way to remove this allergen from fish. In this study, protease gene VSP2V-280 of marine bacteria Virgibacillus sp. SP2 was cloned and expressed. The protease enzyme showed maximum activity at 50°C and pH 10.0. Ca2+ and Cu2+ promoted the enzyme. The enzyme showed good parvalbumin degradation efficiency in fish. Based on the gel analysis, when 0.3 mg/mL of parvalbumin was incubated with protease VSP2V-280 (30 U/mL) containing 1 mM Ca2+ for 3 h, the parvalbumin removal rate reached 97%. The enzyme was further used for parvalbumin removal from Ctenopharyngodon idella, Pelteobagrus fulvidraco, Parabramis pekinensis, and Carassius auratus. The parvalbumin removal rate reached 93% in 4 h at an enzyme dosage of 72 U/mL. The study showed the potential of VSP2V-280 to remove parvalbumin from aquatic products.
Keywords: allergen removal; cloning and expression; enzymatic properties; parvalbumin; protease.
© 2025 The Author(s). Food Science & Nutrition published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Molecular and immunological characterization of grass carp (Ctenopharyngodon idella) parvalbumin Cten i 1: A major fish allergen in Hong Kong.Pediatr Allergy Immunol. 2020 Oct;31(7):792-804. doi: 10.1111/pai.13259. Epub 2020 Jun 26. Pediatr Allergy Immunol. 2020. PMID: 32323392
-
Exploring Fish Parvalbumins through Allergen Names and Gene Identities.Genes (Basel). 2024 Oct 18;15(10):1337. doi: 10.3390/genes15101337. Genes (Basel). 2024. PMID: 39457461 Free PMC article. Review.
-
Characterization of detergent compatible protease from halophilic Virgibacillus sp. CD6.3 Biotech. 2018 Feb;8(2):104. doi: 10.1007/s13205-018-1133-2. Epub 2018 Jan 31. 3 Biotech. 2018. PMID: 29404232 Free PMC article.
-
Quantification of major allergen parvalbumin in 22 species of fish by SDS-PAGE.Food Chem. 2016 Mar 1;194:345-53. doi: 10.1016/j.foodchem.2015.08.037. Epub 2015 Aug 12. Food Chem. 2016. PMID: 26471564
-
Cross-reactive monoclonal antibodies against fish parvalbumins as a tool for studying antigenic similarity of different parvalbumins and analysis of fish extracts.Mol Immunol. 2023 Feb;154:80-95. doi: 10.1016/j.molimm.2023.01.001. Epub 2023 Jan 6. Mol Immunol. 2023. PMID: 36621061
References
LinkOut - more resources
Full Text Sources
Miscellaneous

