This is a preprint.
Crystal structure of MutYX: A novel clusterless adenine DNA glycosylase with a distinct C-terminal domain and 8-Oxoguanine recognition sphere
- PMID: 39803464
- PMCID: PMC11722440
- DOI: 10.1101/2025.01.03.631205
Crystal structure of MutYX: A novel clusterless adenine DNA glycosylase with a distinct C-terminal domain and 8-Oxoguanine recognition sphere
Update in
-
Crystal structure of MutYX: a novel clusterless adenine DNA glycosylase with a distinct C-terminal domain and 8-oxoguanine recognition sphere.Nucleic Acids Res. 2025 Nov 13;53(21):gkaf1127. doi: 10.1093/nar/gkaf1127. Nucleic Acids Res. 2025. PMID: 41273174 Free PMC article.
Abstract
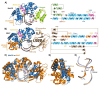
The [4Fe-4S] cluster is an important cofactor of the base excision repair (BER) adenine DNA glycosylase MutY to prevent mutations associated with 8-oxoguanine (OG). Several MutYs lacking the [4Fe-4S] cofactor have been identified. Phylogenetic analysis shows that clusterless MutYs are distributed in two clades suggesting cofactor loss in two independent evolutionary events. Herein, we determined the first crystal structure of a clusterless MutY complexed with DNA. On the basis of the dramatic structural divergence from canonical MutYs, we refer to this as representative of a clusterless MutY subgroup "MutYX". Interestingly, MutYX compensates for the missing [4Fe-4S] cofactor to maintain positioning of catalytic residues by expanding a pre-existing α-helix and acquisition of the new α-helix. Surprisingly, MutYX also acquired a new C-terminal domain that uniquely recognizes OG using residue Gln201 and Arg209. Adenine glycosylase assays and binding affinity measurements indicate that Arg209 is the primary residue responsible to specificity for OG:A lesions, while Gln201 bridges OG and Arg209. Surprisingly, replacement of Arg209 and Gln201 with Ala increases activity toward G:A mismatches. The MutYX structure serves as an example of devolution, capturing structural features required to retain function in the absence of a metal cofactor considered indispensable.
Keywords: Base Excision Repair; DNA glycosylase; Evolution; MutY; [4Fe-4S] cluster.
Figures





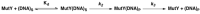
References
-
- Guan Y., Manuel R.C., Arvai A.S., Parikh S.S., Mol C.D., Miller J.H., Lloyd R.S. and Tainer J.A. (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nature Structural and Molecular Biology, 5, 1058. - PubMed
-
- Hinks J.A., Evans M.C., De Miguel Y., Sartori A.A., Jiricny J. and Pearl L.H. (2002) An iron-sulfur cluster in the family 4 uracil-DNA glycosylases. Journal of Biological Chemistry, 277, 16936–16940. - PubMed
-
- Mol C.D., Arvai A.S., Begley T.J., Cunningham R.P. and Tainer J.A. (2002) Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases. Journal of molecular biology, 315, 373–384. - PubMed
-
- Porello S.L., Cannon M.J. and David S.S. (1998) A substrate recognition role for the [4Fe-4S] 2+ cluster of the DNA repair glycosylase MutY. Biochemistry, 37, 6465–6475. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
