This is a preprint.
Pathogenetic mechanisms of muscle-specific ribosomes in dilated cardiomyopathy
- PMID: 39803500
- PMCID: PMC11722222
- DOI: 10.1101/2025.01.02.630345
Pathogenetic mechanisms of muscle-specific ribosomes in dilated cardiomyopathy
Update in
-
Pathogenetic mechanisms of muscle-specific ribosomes in dilated cardiomyopathy.Nat Cardiovasc Res. 2026 Jan;5(1):51-66. doi: 10.1038/s44161-025-00761-8. Epub 2026 Jan 6. Nat Cardiovasc Res. 2026. PMID: 41495453 Free PMC article.
Abstract
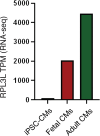
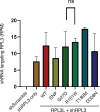
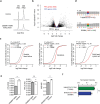
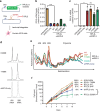
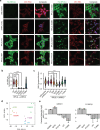
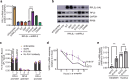
The heart employs a specialized ribosome in its muscle cells to translate genetic information into proteins, a fundamental adaptation with an elusive physiological role1-3. Its significance is underscored by the discovery of neonatal patients suffering from often fatal heart failure caused by severe dilated cardiomyopathy when both copies of the gene RPL3L are mutated4-9. RPL3L is a muscle-specific paralog1-3 of the ubiquitous ribosomal protein L3 (RPL3), which makes the closest contact of any protein to the ribosome's RNA-based catalytic center10. RPL3L-linked heart failure represents the only known human disease associated with tissue-specific ribosomes, yet the underlying pathogenetic mechanisms remain poorly understood. Intriguingly, disease is linked to a large number of mostly missense variants in RPL3L, and RPL3L-knockout resulted in no severe heart defect in either human or mice3, 11-13, challenging the prevailing view that autosomal recessive diseases are caused by loss-of-function mutations. Here, we report three new cases of RPL3L-linked severe neonatal heart failure and present a unifying pathogenetic mechanism by which a large number of variants in the muscle-specific ribosome led to disease. Specifically, affected families often carry one of two recurrent toxic gain-of-function variants alongside a family-specific putative loss-of-function variant. While the non-recurrent variants often trigger partial compensation of RPL3 similar to Rpl3l-knockout mice, both recurrent variants exhibit increased affinity for the RPL3/RPL3L chaperone GRWD114-16 and 60S biogenesis factors, sequester 28S rRNA in the nucleus, disrupt ribosome biogenesis, and trigger severe cellular toxicity that extends beyond the loss of ribosomes. These findings provide critical insights for genetic screening and therapeutic development of neonatal heart failure. Our results suggest that gain-of-toxicity mechanisms may be more prevalent in autosomal recessive diseases, and a combination of gain-of-toxicity and loss-of-function mechanisms could underlie many diseases involving genes with paralogs.
Conflict of interest statement
Declaration of interests X.W. is a member of the Scientific Advisory Board for Epitor Therapeutics. W.K.C. serves on the Board of Directors at Prime Medicine.
Figures



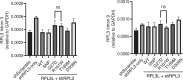

References
-
- Van Raay T.J., Connors T.D., Klinger K.W., Landes G.M. & Burn T.C. A novel ribosomal protein L3-like gene (RPL3L) maps to the autosomal dominant polycystic kidney disease gene region. Genomics 37, 172–176 (1996). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
