This is a preprint.
STAT5B leukemic mutations, altering SH2 tyrosine 665, have opposing impacts on immune gene programs
- PMID: 39803507
- PMCID: PMC11722272
- DOI: 10.1101/2024.12.20.629685
STAT5B leukemic mutations, altering SH2 tyrosine 665, have opposing impacts on immune gene programs
Update in
-
STAT5B leukemic mutations, altering SH2 tyrosine 665, have opposing impacts on immune gene programs.Life Sci Alliance. 2025 Apr 14;8(7):e202503222. doi: 10.26508/lsa.202503222. Print 2025 Jul. Life Sci Alliance. 2025. PMID: 40228864 Free PMC article.
Abstract
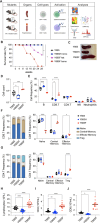
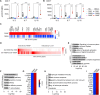
STAT5B is a vital transcription factor for lymphocytes. Here, function of two STAT5B mutations from human T cell leukemias: one substituting tyrosine 665 with phenylalanine (STAT5BY665F), the other with histidine (STAT5BY665H) was interrogated. In silico modeling predicted divergent energetic effects on homodimerization with a range of pathogenicity. In primary T cells in vitro STAT5BY665F showed gain-of-function while STAT5BY665H demonstrated loss-of-function. Introducing the mutation into the mouse genome illustrated that the gain-of-function Stat5b Y665F mutation resulted in accumulation of CD8+ effector and memory and CD4+ regulatory T-cells, altering CD8+/CD4+ ratios. In contrast, STAT5BY665H 'knock-in' mice showed diminished CD8+ effector and memory and CD4+ regulatory T cells. In contrast to wild-type STAT5, the STAT5BY665F variant displayed greater STAT5 phosphorylation, DNA binding and transcriptional activity following cytokine activation while the STAT5BY665H variant resembled a null. The work exemplifies how joining in silico and in vivo studies of single nucleotides deepens our understanding of disease-associated variants, revealing structural determinants of altered function, defining mechanistic roles, and, specifically here, identifying a gain-of function variant that does not directly induce hematopoietic malignancy.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
