Cardiac-targeted delivery of a novel Drp1 inhibitor for acute cardioprotection
- PMID: 39803589
- PMCID: PMC11708310
- DOI: 10.1016/j.jmccpl.2024.100085
Cardiac-targeted delivery of a novel Drp1 inhibitor for acute cardioprotection
Abstract
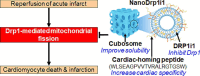
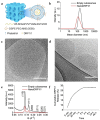
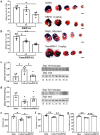
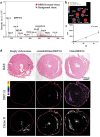
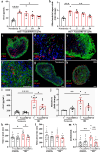
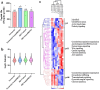
Dynamin-related protein 1 (Drp1) is a mitochondrial fission protein and a viable target for cardioprotection against myocardial ischaemia-reperfusion injury. Here, we reported a novel Drp1 inhibitor (DRP1i1), delivered using a cardiac-targeted nanoparticle drug delivery system, as a more effective approach for achieving acute cardioprotection. DRP1i1 was encapsulated in cubosome nanoparticles with conjugated cardiac-homing peptides (NanoDRP1i1) and the encapsulation efficiency was 99.3 ± 0.1 %. In vivo, following acute myocardial ischaemia-reperfusion injury in mice, NanoDRP1i1 significantly reduced infarct size and serine-616 phosphorylation of Drp1, and restored cardiomyocyte mitochondrial size to that of sham group. Imaging by mass spectrometry revealed higher accumulation of DRP1i1 in the heart tissue when delivered as NanoDRP1i1. In human cardiac organoids subjected to simulated ischaemia-reperfusion injury, treatment with NanoDRP1i1 at reperfusion significantly reduced cardiac cell death, contractile dysfunction, and mitochondrial superoxide levels. Following NanoDRP1i1 treatment, cardiac organoid proteomics further confirmed reprogramming of contractile dysfunction markers and enrichment of the mitochondrial protein network, cytoskeletal and metabolic regulation networks when compared to the simulated injury group. These proteins included known cardioprotective regulators identified in human organoids and in vivo murine studies following ischaemia-reperfusion injury. DRP1i1 is a promising tool compound to study Drp1-mediated mitochondrial fission and exhibits promising therapeutic potential for acute cardioprotection, especially when delivered using the cardiac-targeted cubosome nanoparticles.
Keywords: Cardiac organoids; Cubosome; Dynamin-related protein 1; Mitochondria; Myocardial ischaemia-reperfusion injury.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Heidenreich P.A., Bozkurt B., Aguilar D., Allen L.A., Byun J.J., Colvin M.M., et al. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: executive summary: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation. 2022;145(18) - PubMed
-
- Hausenloy D.J., Yellon D.M. Targeting myocardial reperfusion injury--the search continues. N Engl J Med. 2015;373(11):1073–1075. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

