Microcephaly protein ANKLE2 promotes Zika virus replication
- PMID: 39804047
- PMCID: PMC11796389
- DOI: 10.1128/mbio.02683-24
Microcephaly protein ANKLE2 promotes Zika virus replication
Abstract
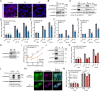
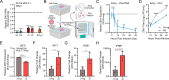
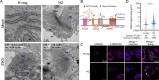
Orthoflaviviruses are positive-sense single-stranded RNA viruses that hijack host proteins to promote their own replication. Zika virus (ZIKV) is infamous among orthoflaviviruses for its association with severe congenital birth defects, notably microcephaly. We previously mapped ZIKV-host protein interactions and identified the interaction between ZIKV non-structural protein 4A (NS4A) and host microcephaly protein ankyrin repeat and LEM domain-containing 2 (ANKLE2). Using a fruit fly model, we showed that NS4A induced microcephaly in an ANKLE2-dependent manner. Here, we explore the role of ANKLE2 in ZIKV replication to understand the biological significance of the interaction from a viral perspective. We observe that ANKLE2 localization is drastically shifted to sites of NS4A accumulation during infection and that knockout of ANKLE2 reduces ZIKV replication in multiple human cell lines. This decrease in virus replication is coupled with a moderate increase in innate immune activation. Using microscopy, we observe dysregulated formation of virus-induced endoplasmic reticulum rearrangements in ANKLE2 knockout cells. Knockdown of the ANKLE2 ortholog in Aedes aegypti cells also decreases virus replication, suggesting ANKLE2 is a beneficial replication factor across hosts. Finally, we show that NS4A from four other orthoflaviviruses physically interacts with ANKLE2 and is also beneficial to their replication. Thus, ANKLE2 likely promotes orthoflavivirus replication by regulating membrane rearrangements that serve to accelerate viral genome replication and protect viral dsRNA from immune detection. Taken together with our previous results, our findings indicate that ZIKV and other orthoflaviviruses hijack ANKLE2 for a conserved role in replication, and this drives unique pathogenesis for ZIKV since ANKLE2 has essential roles in developing tissues.IMPORTANCEZIKV is a major concern due to its association with birth defects, including microcephaly. We previously identified a physical interaction between ZIKV NS4A and host microcephaly protein ANKLE2. Mutations in ANKLE2 cause congenital microcephaly, and NS4A induces microcephaly in an ANKLE2-dependent manner. Here, we establish the role of ANKLE2 in ZIKV replication. Depletion of ANKLE2 from cells significantly reduces ZIKV replication and disrupts virus-induced membrane rearrangements. ANKLE2's ability to promote ZIKV replication is conserved in mosquito cells and for other related mosquito-borne orthoflaviviruses. Our data point to an overall model in which ANKLE2 regulates virus-induced membrane rearrangements to accelerate orthoflavivirus replication and avoid immune detection. However, ANKLE2's unique role in ZIKV NS4A-induced microcephaly is a consequence of ZIKV infection of important developing tissues in which ANKLE2 has essential roles.
Keywords: ANKLE2; NS4A; Zika virus; microcephaly; orthoflavivirus; virus-host interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

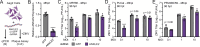

References
-
- Cortese M, Goellner S, Acosta EG, Neufeldt CJ, Oleksiuk O, Lampe M, Haselmann U, Funaya C, Schieber N, Ronchi P, Schorb M, Pruunsild P, Schwab Y, Chatel-Chaix L, Ruggieri A, Bartenschlager R. 2017. Ultrastructural characterization of Zika virus replication factories. Cell Rep 18:2113–2123. doi: 10.1016/j.celrep.2017.02.014 - DOI - PMC - PubMed
-
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- S10 OD026702/OD/NIH HHS/United States
- R01 AI170857/AI/NIAID NIH HHS/United States
- T32 AI060555/AI/NIAID NIH HHS/United States
- T34 GM136469/GM/NIGMS NIH HHS/United States
- Summer Undergraduate Research Program Fellowship/UC | UCD | UC Davis College of Biological Sciences (CBS)
- R21 AI168716/AI/NIAID NIH HHS/United States
- 2T32AI060555-16/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- 5T32GM7377-42/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- R56 AI170857/AI/NIAID NIH HHS/United States
- W. M. Keck Foundation (WMKF)
- R56AI170857,R01AI170857,R21AI168716/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- T34GM136469/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- T32 GM007377/GM/NIGMS NIH HHS/United States
- S10 RR019266/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
