PHIStruct: improving phage-host interaction prediction at low sequence similarity settings using structure-aware protein embeddings
- PMID: 39804673
- PMCID: PMC11783280
- DOI: 10.1093/bioinformatics/btaf016
PHIStruct: improving phage-host interaction prediction at low sequence similarity settings using structure-aware protein embeddings
Abstract
Motivation: Recent computational approaches for predicting phage-host interaction have explored the use of sequence-only protein language models to produce embeddings of phage proteins without manual feature engineering. However, these embeddings do not directly capture protein structure information and structure-informed signals related to host specificity.
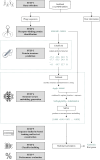
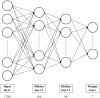
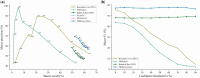
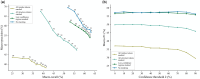
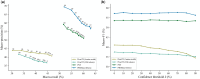
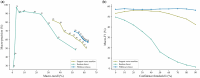
Results: We present PHIStruct, a multilayer perceptron that takes in structure-aware embeddings of receptor-binding proteins, generated via the structure-aware protein language model SaProt, and then predicts the host from among the ESKAPEE genera. Compared against recent tools, PHIStruct exhibits the best balance of precision and recall, with the highest and most stable F1 score across a wide range of confidence thresholds and sequence similarity settings. The margin in performance is most pronounced when the sequence similarity between the training and test sets drops below 40%, wherein, at a relatively high-confidence threshold of above 50%, PHIStruct presents a 7%-9% increase in class-averaged F1 over machine learning tools that do not directly incorporate structure information, as well as a 5%-6% increase over BLASTp.
Availability and implementation: The data and source code for our experiments and analyses are available at https://github.com/bioinfodlsu/PHIStruct.
© The Author(s) 2025. Published by Oxford University Press.
Figures


References
-
- Antimicrobial resistance surveillance in Europe 2023 - 2021 data. Stockholm: European Centre for Disease Prevention and Control and World Health Organization, 2023.
-
- Badam S, Rao S. Harnessing genome representation learning for decoding phage–host interactions. bioRxiv, 2024, preprint: not peer reviewed.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

