Exercise intensity and training alter the innate immune cell type and chromosomal origins of circulating cell-free DNA in humans
- PMID: 39805013
- PMCID: PMC11761974
- DOI: 10.1073/pnas.2406954122
Exercise intensity and training alter the innate immune cell type and chromosomal origins of circulating cell-free DNA in humans
Abstract
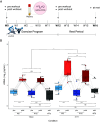
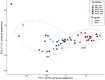
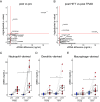
Exercising regularly promotes health, but these benefits are complicated by acute inflammation induced by exercise. A potential source of inflammation is cell-free DNA (cfDNA), yet the cellular origins, molecular causes, and immune system interactions of exercise-induced cfDNA are unclear. To study these, 10 healthy individuals were randomized to a 12-wk exercise program of either high-intensity tactical training (HITT) or traditional moderate-intensity training (TRAD). Blood plasma was collected pre- and postexercise at weeks 0 and 12 and after 4 wk of detraining upon program completion. Whole-genome enzymatic methylation sequencing (EM-seq) with cell-type proportion deconvolution was applied to cfDNA obtained from the 50 plasma samples and paired to concentration measurements for 90 circulating cytokines. Acute exercise increased the release of cfDNA from neutrophils, dendritic cells (DCs), and macrophages proportional to exercise intensity. Exercise training reduced cfDNA released in HITT participants but not TRAD and from DCs and macrophages but not neutrophils. For most participants, training lowered mitochondrial cfDNA at rest, even after detraining. Using a sequencing analysis approach we developed, we concluded that rapid ETosis, a process of cell death where cells release DNA extracellular traps, was the likely source of cfDNA, demonstrated by enrichment of nuclear DNA. Further, several cytokines were induced by acute exercise, such as IL-6, IL-10, and IL-16, and training attenuated the induction of only IL-6 and IL-17F. Cytokine levels were not associated with cfDNA induction, suggesting that these cytokines are not the main cause of exercise-induced cfDNA. Overall, exercise intensity and training modulated cfDNA release and cytokine responses, contributing to the anti-inflammatory effects of regular exercise.
Keywords: DNA methylation; ETosis; circulating cell-free DNA; exercise biology; extracellular traps.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Mandel P., Metais P., Nuclear acids in human blood plasma. C. R. Seances Soc. Biol. Fil. 142, 241–243 (1948). - PubMed
-
- De Borre M., et al. , Cell-free DNA methylome analysis for early preeclampsia prediction. Nat. Med. 29, 2206–2215 (2023). - PubMed
-
- Poon L. L. M., Leung T. N., Lau T. K., Chow K. C. K., Lo Y. M. D., Differential DNA methylation between fetus and mother as a strategy for detecting fetal DNA in maternal plasma. Clin. Chem. 48, 35–41 (2002). - PubMed
-
- Nawroz H., Koch W., Anker P., Stroun M., Sidransky D., Microsatellite alterations in serum DNA of head and neck cancer patients. Nat. Med. 2, 1035–1037 (1996). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

