Oral obeldesivir provides postexposure protection against Marburg virus in nonhuman primates
- PMID: 39805309
- PMCID: PMC12003170
- DOI: 10.1038/s41591-025-03496-y
Oral obeldesivir provides postexposure protection against Marburg virus in nonhuman primates
Abstract
The recent outbreak of Marburg virus (MARV) in Rwanda underscores the need for effective countermeasures against this highly fatal pathogen, with case fatality rates reaching 90%. Currently, no vaccines or approved treatments exist for MARV infection, distinguishing it from related viruses such as Ebola. Our study demonstrates that the oral drug obeldesivir (ODV), a nucleoside analog prodrug, shows promising antiviral activity against filoviruses in vitro and offers significant protection in animal models. Here with cynomolgus macaques (n = 6), a 10 day regimen of once-daily ODV, initiated 24 h after exposure, provided 80% protection against a thousandfold lethal MARV challenge, delaying viral replication and disease onset. Transcriptome analysis revealed that early adaptive responses correlated with successful outcomes. Compared with intravenous options, oral antivirals such as ODV offer logistical advantages in outbreak settings, enabling easier administration and broader contact coverage. Our findings support the potential of ODV as a broad-spectrum, oral postexposure prophylaxis for filoviruses.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: T.C., A.-Q.N., D.B., R.B., M.S.V. and V.C.C. are paid employees of Gilead Sciences, Inc. and may own company stock. The other authors declare no competing interests.
Figures
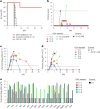
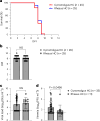



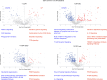
References
-
- Feldmann, H., Sanchez, A. & Geisbert, T. W. in Fields Virology Vol. 1 (eds Knipe, D. M. & Howley, P. M.) Ch. 32, 923–956 (Lippincott Williams & Wilkins, 2013).
-
- Feldmann, H., Sprecher, A. & Geisbert, T. W. Ebola. N. Engl. J. Med.382, 1832–1842 (2020). - PubMed
-
- Bausch, D. G. et al. Marburg hemorrhagic fever associated with multiple genetic lineages of virus. N. Engl. J. Med.355, 909–919 (2006). - PubMed
-
- World Health Organization. Marburg virus disease—Rwanda https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON537 (2024).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

