Longitudinal serum proteome mapping reveals biomarkers for healthy ageing and related cardiometabolic diseases
- PMID: 39805987
- PMCID: PMC11774760
- DOI: 10.1038/s42255-024-01185-7
Longitudinal serum proteome mapping reveals biomarkers for healthy ageing and related cardiometabolic diseases
Abstract
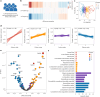
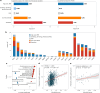
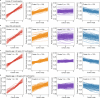
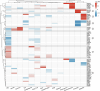
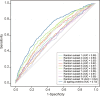
The blood proteome contains biomarkers of ageing and age-associated diseases, but such markers are rarely validated longitudinally. Here we map the longitudinal proteome in 7,565 serum samples from a cohort of 3,796 middle-aged and elderly adults across three time points over a 9-year follow-up period. We pinpoint 86 ageing-related proteins that exhibit signatures associated with 32 clinical traits and the incidence of 14 major ageing-related chronic diseases. Leveraging a machine-learning model, we pick 22 of these proteins to generate a proteomic healthy ageing score (PHAS), capable of predicting the incidence of cardiometabolic diseases. We further identify the gut microbiota as a modifiable factor influencing the PHAS. Our data constitute a valuable resource and offer useful insights into the roles of serum proteins in ageing and age-associated cardiometabolic diseases, providing potential targets for intervention with therapeutics to promote healthy ageing.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: T.G. is a shareholder of Westlake Omics. The other authors declare no competing interests.
Figures










References
-
- López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell186, 243–278 (2023). - PubMed
MeSH terms
Substances
Grants and funding
- 82073529/National Natural Science Foundation of China (National Science Foundation of China)
- 82103828/National Natural Science Foundation of China (National Science Foundation of China)
- 82103826/National Natural Science Foundation of China (National Science Foundation of China)
- 82204161/National Natural Science Foundation of China (National Science Foundation of China)
- 82073546/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical

