Ultrasensitive ctDNA detection for preoperative disease stratification in early-stage lung adenocarcinoma
- PMID: 39806071
- PMCID: PMC11750713
- DOI: 10.1038/s41591-024-03216-y
Ultrasensitive ctDNA detection for preoperative disease stratification in early-stage lung adenocarcinoma
Abstract
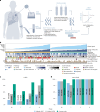
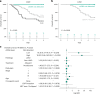
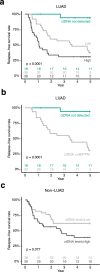
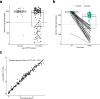
Circulating tumor DNA (ctDNA) detection can predict clinical risk in early-stage tumors. However, clinical applications are constrained by the sensitivity of clinically validated ctDNA detection approaches. NeXT Personal is a whole-genome-based, tumor-informed platform that has been analytically validated for ultrasensitive ctDNA detection at 1-3 ppm of ctDNA with 99.9% specificity. Through an analysis of 171 patients with early-stage lung cancer from the TRACERx study, we detected ctDNA pre-operatively within 81% of patients with lung adenocarcinoma (LUAD), including 53% of those with pathological TNM (pTNM) stage I disease. ctDNA predicted worse clinical outcome, and patients with LUAD with <80 ppm preoperative ctDNA levels (the 95% limit of detection of a ctDNA detection approach previously published in TRACERx) experienced reduced overall survival compared with ctDNA-negative patients with LUAD. Although prospective studies are needed to confirm the clinical utility of the assay, these data show that our approach has the potential to improve disease stratification in early-stage LUADs.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.A.B. has consulted for Achilles Therapeutics. D.A.M. reports receiving speaker fees from AstraZeneca, Eli Lilly, Bristol Myers Squibb and Takeda, consultancy fees from AstraZeneca, Thermo Fisher, Takeda, Amgen, Janssen, MIM Software, Bristol Myers Squibb and Eli Lilly and has received educational support from Takeda and Amgen. M.J.-H. has consulted for, and is a member of, the Achilles Therapeutics scientific advisory board (SAB) and steering committee, and has received speaker honoraria from Pfizer, Astex Pharmaceuticals and the Oslo Cancer Cluster. N.M. has received consultancy fees and has stock options in Achilles Therapeutics. C.S. acknowledges grants from AstraZeneca, Boehringer-Ingelheim, Bristol Myers Squibb, Pfizer, Roche-Ventana, Invitae (previously Archer Dx Inc, collaboration in minimal residual disease sequencing technologies), Ono Pharmaceutical and Personalis. He is chief investigator for the AZ MeRmaiD 1 and 2 clinical trials and is the steering committee chair. He is also co-chief investigator of the NHS Galleri trial funded by GRAIL and a paid member of GRAIL’s SAB. He receives consultant fees from Achilles Therapeutics (and is a SAB member), Bicycle Therapeutics (and is a SAB member), Genentech, Medicxi, China Innovation Centre of Roche (formerly Roche Innovation Centre – Shanghai, Metabomed (until July 2022)), Relay Therapeutics (and is a SAB member), Saga Diagnostics (and is a SAB member) and the Sarah Cannon Research Institute. C.S has received honoraria from Amgen, AstraZeneca, Bristol Myers Squibb, GSK, Illumina, MSD, Novartis, Pfizer and Roche-Ventana. C.S. has previously held stock options in Apogen Biotechnologies and GRAIL, and currently has stock options in Bicycle Therapeutics and Relay Therapeutics, and has stock and is a co-founder of Achilles Therapeutics. G.B., C.W.A., S.M.B, R.C., J.H., B.L., J.L., F.C.P.N., J.N., R.M.P. and R.O.C. are employees and stockholders of Personalis. A.M.F. is listed as a co-inventor on a patent application to determine methods and systems for tumor monitoring (PCT/EP2022/077987; ‘Methods and systems for tumour monitoring’). S.V. is a co-inventor on a patent for methods for detecting molecules in a sample (US patent no. 10578620; ‘Methods for detecting molecules in a sample’). M.J.-H. is listed as a co-inventor on a European patent application related to methods to detect lung cancer (PCT/US2017/028013; ‘Methods for lung cancer detection’); this patent has been licensed to commercial entities and, under terms of employment, M.J.-H. is due a share of any revenue generated from such license(s). N.M. holds European patents related to targeting neoantigens (PCT/EP2016/059401; ‘Method for treating cancer’), identifying patient response to immune checkpoint blockade (PCT/EP2016/071471; ‘“Immune checkpoint intervention” in cancer’), determining HLA LOH (PCT/GB2018/052004; ‘Analysis of HLA alleles in tumours and the uses thereof’), and predicting survival rates of patients with cancer (PCT/GB2020/050221; ‘Method of predicting survival rates for cancer patients’). C.S declares a patent application for methods to lung cancer (PCT/US2017/028013); targeting neoantigens (PCT/EP2016/059401); identifying patent response to immune checkpoint blockade (PCT/EP2016/071471); methods for lung cancer detection (US20190106751A1); identifying patients who respond to cancer treatment (PCT/GB2018/051912); determining HLA LOH (PCT/GB2018/052004); predicting survival rates of patients with cancer (PCT/GB2020/050221); and methods and systems for tumor monitoring (PCT/EP2022/077987). C.S. is an inventor on a European patent application (PCT/GB2017/053289) related to assay technology to detect tumor recurrence. This patent has been licensed to a commercial entity, and under their terms of employment, C.S is due a revenue share of any revenue generated from such license(s). The other authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

