Scale-free Spatio-temporal Correlations in Conformational Fluctuations of Intrinsically Disordered Proteins
- PMID: 39807013
- PMCID: PMC11884614
- DOI: 10.1002/advs.202412989
Scale-free Spatio-temporal Correlations in Conformational Fluctuations of Intrinsically Disordered Proteins
Abstract
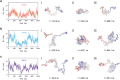
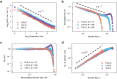
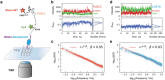
The self-assembly of intrinsically disordered proteins (IDPs) into condensed phases and the formation of membrane-less organelles (MLOs) can be considered as the phenomenon of collective behavior. The conformational dynamics of IDPs are essential for their interactions and the formation of a condensed phase. From a physical perspective, collective behavior and the emergence of phase are associated with long-range correlations. Here the conformational dynamics of IDPs and the correlations therein are analyzed, using µs-scale atomistic molecular dynamics (MD) simulations and single-molecule Förster resonance energy transfer (smFRET) experiments. The existence of typical scale-free spatio-temporal correlations in IDP conformational fluctuations is demonstrated. Their conformational evolutions exhibit "1/f noise" power spectra and are accompanied by the appearance of residue domains following a power-law size distribution. Additionally, the motions of residues present scale-free behavioral correlation. These scale-free correlations resemble those in physical systems near critical points, suggesting that IDPs are poised at a critical state. Therefore, IDPs can effectively respond to finite differences in sequence compositions and engender considerable structural heterogeneity which is beneficial for IDP interactions and phase formation.
Keywords: collective behavior; conformational dynamics; critical phenomena; intrinsically disordered proteins; scale‐free correlation.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
