QOT: Quantized Optimal Transport for sample-level distance matrix in single-cell omics
- PMID: 39808114
- PMCID: PMC11962597
- DOI: 10.1093/bib/bbae713
QOT: Quantized Optimal Transport for sample-level distance matrix in single-cell omics
Abstract
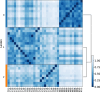
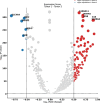
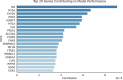
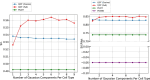
Single-cell technologies have enabled the high-dimensional characterization of cell populations at an unprecedented scale. The innate complexity and increasing volume of data pose significant computational and analytical challenges, especially in comparative studies delineating cellular architectures across various biological conditions (i.e. generation of sample-level distance matrices). Optimal Transport is a mathematical tool that captures the intrinsic structure of data geometrically and has been applied to many bioinformatics tasks. In this paper, we propose QOT (Quantized Optimal Transport), a new method enabling efficient computation of sample-level distance matrix from large-scale single-cell omics data through a quantization step. We apply our algorithm to real-world single-cell genomics and pathomics datasets, aiming to extrapolate cell-level insights to inform sample-level categorizations. Our empirical study shows that QOT outperforms existing two OT-based algorithms in accuracy and robustness when obtaining a distance matrix from high throughput single-cell measures at the sample level. Moreover, the sample level distance matrix could be used in the downstream analysis (i.e. uncover the trajectory of disease progression), highlighting its usage in biomedical informatics and data science.
Keywords: Gaussian Mixture Model; Wasserstein distance; optimal transport; quantization; single-cell genomics.
© The Author(s) 2025. Published by Oxford University Press.
Figures



Update of
-
QOT: Efficient Computation of Sample Level Distance Matrix from Single-Cell Omics Data through Quantized Optimal Transport.bioRxiv [Preprint]. 2024 Feb 6:2024.02.06.578032. doi: 10.1101/2024.02.06.578032. bioRxiv. 2024. Update in: Brief Bioinform. 2024 Nov 22;26(1):bbae713. doi: 10.1093/bib/bbae713. PMID: 38370767 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

