Naturally occurring variation in a cytochrome P450 modifies thiabendazole responses independently of beta-tubulin
- PMID: 39808673
- PMCID: PMC11771912
- DOI: 10.1371/journal.ppat.1012602
Naturally occurring variation in a cytochrome P450 modifies thiabendazole responses independently of beta-tubulin
Abstract
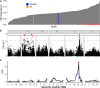
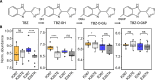
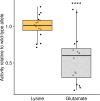
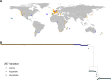
Widespread anthelmintic resistance has complicated the management of parasitic nematodes. Resistance to the benzimidazole (BZ) drug class is nearly ubiquitous in many species and is associated with mutations in beta-tubulin genes. However, mutations in beta-tubulin alone do not fully explain all BZ resistance. We performed a genome-wide association study using a genetically diverse panel of Caenorhabditis elegans strains to identify loci that contribute to resistance to the BZ drug thiabendazole (TBZ). We identified a quantitative trait locus (QTL) on chromosome V independent of all beta-tubulin genes and overlapping with two promising candidate genes, the cytochrome P450 gene cyp-35D1 and the nuclear hormone receptor nhr-176. Both genes were previously demonstrated to play a role in TBZ metabolism. NHR-176 binds TBZ and induces the expression of CYP-35D1, which metabolizes TBZ. We generated single gene deletions of cyp-35D1 and nhr-176 and found that both genes play a role in TBZ response. A predicted high-impact lysine-to-glutamate substitution at position 267 (K267E) in CYP-35D1 was identified in a sensitive strain, and reciprocal allele replacement strains in different genetic backgrounds were used to show that the lysine allele conferred increased TBZ resistance. Using competitive fitness assays, we found that neither allele was deleterious, but the lysine allele was selected in the presence of TBZ. Additionally, we found that the lysine allele significantly increased the rate of TBZ metabolism compared to the glutamate allele. Moreover, yeast expression assays showed that the lysine version of CYP-35D1 had twice the enzymatic activity of the glutamate allele. To connect our results to parasitic nematodes, we analyzed four Haemonchus contortus cytochrome P450 orthologs but did not find variation at the 267 position in fenbendazole-resistant populations. Overall, we confirmed that variation in this cytochrome P450 gene is the first locus independent of beta-tubulin to play a role in BZ resistance.
Copyright: © 2025 Collins et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

