Identification and validation of a novel defined stress granule-related gene signature for predicting the prognosis of ovarian cancer via bioinformatics analysis
- PMID: 39809219
- PMCID: PMC11596697
- DOI: 10.1097/MD.0000000000040608
Identification and validation of a novel defined stress granule-related gene signature for predicting the prognosis of ovarian cancer via bioinformatics analysis
Abstract
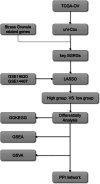
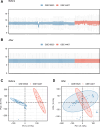
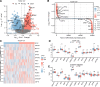
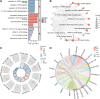
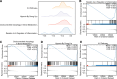
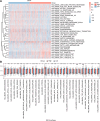
Ovarian cancer (OC) is a malignant gynecological cancer with an extremely poor prognosis. Stress granules (SGs) are non-membrane organelles that respond to stressors; however, the correlation between SG-related genes and the prognosis of OC remains unclear. This systematic analysis aimed to determine the expression levels of SG-related genes between high- and low-risk groups of patients with OC and to explore the prognostic value of these genes. RNA-sequencing data and clinical information from GSE18520 and GSE14407 in the Gene Expression Omnibus (GEO) and ovarian plasmacytoma adenocarcinoma in The Cancer Genome Atlas (TCGA) were downloaded. SG-related genes were obtained from GeneCards, the Molecular Signatures Database, and the literature. First, 13 SG-related genes were identified in the prognostic model using least absolute shrinkage and selection operator (LASSO) Cox regression. The prognostic value of each SG-related gene for survival and its relationship with clinical characteristics were evaluated. Next, we performed a functional enrichment analysis of SG-related genes. The protein-protein interactions (PPI) of SG-related genes were visualized using Cytoscape with STRING. According to the median risk score from the LASSO Cox regression, a 13-gene signature was created. All patients with OC in TCGA cohort and GEO datasets were classified into high- and low-risk groups. Five SG-related genes were differentially expressed between the high- and low-risk OC groups in the GEO datasets. The 13 SG-related genes were related to several important oncogenic pathways (TNF-α signaling, PI3K-AKT-mTOR signaling, and WNT-β-catenin signaling) and several cellular components (cytoplasmic stress granule, cytoplasmic ribonucleoprotein granule, and ribonucleoprotein granule). The PPI network identified 11 hub genes with the strongest interactions with ELAVL1. These findings indicate that SG-related genes (DNAJA1, ELAVL1, FBL, GRB7, MOV10, PABPC3, PCBP2, PFN1, RFC4, SYNCRIP, USP10, ZFP36, and ZFP36L1) can be used to predict OC prognosis.
Copyright © 2024 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors have no conflict of interest to disclose.
Figures




References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

