Parallel genetic adaptation amid a background of changing effective population sizes in divergent yellow perch (Perca flavescens) populations
- PMID: 39809312
- PMCID: PMC11732403
- DOI: 10.1098/rspb.2024.2339
Parallel genetic adaptation amid a background of changing effective population sizes in divergent yellow perch (Perca flavescens) populations
Abstract
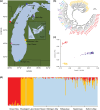
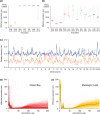
Aquatic ecosystems are highly dynamic environments vulnerable to natural and anthropogenic disturbances. High-economic-value fisheries are one of many ecosystem services affected by these disturbances, and it is critical to accurately characterize the genetic diversity and effective population sizes of valuable fish stocks through time. We used genome-wide data to reconstruct the demographic histories of economically important yellow perch (Perca flavescens) populations. In two isolated and genetically divergent populations, we provide independent evidence for simultaneous increases in effective population sizes over both historic and contemporary time scales including negative genome-wide estimates of Tajima's D, 3.1 times more single nucleotide polymorphisms than adjacent populations, and contemporary effective population sizes that have increased 10- and 47-fold from their minimum, respectively. The excess of segregating sites and negative Tajima's D values probably arose from mutations accompanying historic population expansions with insufficient time for purifying selection, whereas linkage disequilibrium-based estimates of Ne also suggest contemporary increases that may have been driven by reduced fishing pressure or environmental remediation. We also identified parallel, genetic adaptation to reduced visual clarity in the same two habitats. These results suggest that the synchrony of key ecological and evolutionary processes can drive parallel demographic and evolutionary trajectories across independent populations.
Keywords: demographic history; effective population size; fisheries; genetic adaptation; genetic diversity; parallel evolution.
Conflict of interest statement
We declare we have no competing interests.
Figures


Similar articles
-
Waterscape genetics of the yellow perch (Perca flavescens): patterns across large connected ecosystems and isolated relict populations.Mol Ecol. 2012 Dec;21(23):5795-826. doi: 10.1111/mec.12044. Epub 2012 Oct 18. Mol Ecol. 2012. PMID: 23078285
-
Evolutionary ecotoxicology of wild yellow perch (Perca flavescens) populations chronically exposed to a polymetallic gradient.Aquat Toxicol. 2008 Jan 20;86(1):76-90. doi: 10.1016/j.aquatox.2007.10.003. Epub 2007 Oct 12. Aquat Toxicol. 2008. PMID: 18031837
-
Genetic structure and phenotypic plasticity of yellow perch (Perca flavescens) populations influenced by habitat, predation, and contamination gradients.Integr Environ Assess Manag. 2008 Apr;4(2):264-6. Integr Environ Assess Manag. 2008. PMID: 18494119 No abstract available.
-
Comparative transcriptome analysis reveals potential evolutionary differences in adaptation of temperature and body shape among four Percidae species.PLoS One. 2019 May 7;14(5):e0215933. doi: 10.1371/journal.pone.0215933. eCollection 2019. PLoS One. 2019. PMID: 31063465 Free PMC article.
-
Balancing selection on the number of repeats in the ribosomal intergenic spacer present in naturally occurring yellow perch (Perca flavescens) populations.Genome. 2018 Jan;61(1):1-6. doi: 10.1139/gen-2017-0061. Epub 2017 Sep 26. Genome. 2018. PMID: 28950069
References
-
- Hedgecock D, Pudovkin AI. 2011. Sweepstakes reproductive success in highly fecund marine fish and shellfish: a review and commentary. Bull. Mar. Sci. 87, 971–1002. (10.5343/bms.2010.1051) - DOI
-
- Hedgecock D. 1994. Does variance in reproductive success limit effective population sizes of marine organisms? In Genetics and evolution of aquatic organisms (ed. Beaumont A), pp. 122–134. New York, NY: Springer.
-
- Hoarau G, Boon E, Jongma DN, Ferber S, Palsson J, Van der Veer HW, Rijnsdorp AD, Stam WT, Olsen JL. 2005. Low effective population size and evidence for inbreeding in an overexploited flatfish, plaice (Pleuronectes platessa L.). Proc. R. Soc. B 272, 497–503. (10.1098/rspb.2004.2963) - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

