Optimizing the NGS-based discrimination of multiple lung cancers from the perspective of evolution
- PMID: 39809905
- PMCID: PMC11733135
- DOI: 10.1038/s41698-024-00786-5
Optimizing the NGS-based discrimination of multiple lung cancers from the perspective of evolution
Abstract
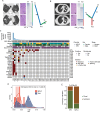
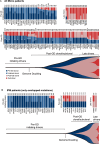
Next-generation sequencing (NGS) offers a promising approach for differentiating multiple primary lung cancers (MPLC) from intrapulmonary metastasis (IPM), though panel selection and clonal interpretation remain challenging. Whole-exome sequencing (WES) data from 80 lung cancer samples were utilized to simulate MPLC and IPM, with various sequenced panels constructed through gene subsampling. Two clonal interpretation approaches primarily applied in clinical practice, MoleA (based on shared mutation comparison) and MoleB (based on probability calculation), were subsequently evaluated. ROC analysis highlighted MoleB's superior performance, especially with the NCCNplus panel (AUC = 0.950 ± 0.002) and pancancer MoleA (AUC = 0.792 ± 0.004). In two independent cohorts (WES cohort, N = 42 and non-WES cohort, N = 94), NGS-based methodologies effectively stratified disease-free survival, with NCCNplus MoleB further predicting prognosis. Phylogenetic analysis further revealed evolutionary distinctions between MPLC and IPM, establishing an optimized NGS-based framework for differentiating multiple lung cancers.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
LinkOut - more resources
Full Text Sources

