The complete mitochondrial genome of Gyrodactylus pseudorasborae (Platyhelminthes: Monogenea) with a phylogeny of Gyrodactylidae parasites
- PMID: 39810079
- PMCID: PMC11730495
- DOI: 10.1186/s12864-025-11225-5
The complete mitochondrial genome of Gyrodactylus pseudorasborae (Platyhelminthes: Monogenea) with a phylogeny of Gyrodactylidae parasites
Abstract
Background: Gyrodactylus von Nordmann, 1832, a genus of viviparous parasites within the family Gyrodactylidae, contains one of the largest nominal species in the world. Gyrodactylus pseudorasborae Ondračková, Seifertová & Tkachenko, 2023 widely distributed in Europe and China, although its mitochondrial genome remains unclear. This study aims to sequence the mitogenome of G.pseudorasborae and clarify its phylogenetic relationship within the Gyrodactylidea. The mitochondrial genome of G. pseudorasborae was amplified in six parts from a single parasite, sequenced using primer walking, annotated and analyzed using bioinformatic tools.
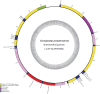
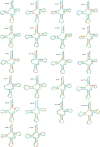
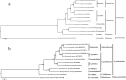
Results: The mitochondrial genome of G. pseudorasborae is 14,189 bp in length, containing 12 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs), and two major non-coding regions (NCR: NC1 and NC2). The overall A + T content of the mitogenome is 73.1%, a medium content compared with all reported mitochondrial genomes of monogeneans. The mitogenome of G. pseudorasborae presents a clear bias in nucleotide composition with a negative AT skew and a positive GC skew except for NCR. All tRNAs have the typical cloverleaf secondary structure except for tRNACys, tRNASer1, and tRNASer2, which lack the dihydrouridine (DHU) arm. Furthermore, one repetitive non-coding region of 32 bp repeats occurred in the NC1 region with poly-T stretch, stem-loop structure, and TAn motif. The gene order is identical to the mitochondrial genomes reported from other Gyrodactylus species except Gyrodactylus nyanzae Paperna, 1973 and Gyrodactylus sp. FZ-2021. Phylogenetic analyses show that G. pseudorasborae and Gyrodactylus parvae You, Easy & Cone, 2008 cluster together with high nodal support based on 12 PCGs sequences and amino acid sequences, Gyrodactylidae forms independent monophyletic clade within Gyrodactylidea.
Conclusion: Both the mitochondrial genome and phylogenetic analyses support G. pseudorasborae is a member of the genus Gyrodactylus and Gyrodactylidae forms an independent monophyletic clade within Gyrodactylidea. Furthermore, the mitochondrial genome of G. pseudorasborae is the shortest in the Gyrodactylidea species compared with size differences in NCR.
Keywords: Gyrodactylus pseudorasborae; Pseudorasbora parva; China; Gyrodactylidea; Mitochondrial genome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the Animal Care and Use Committee of Guilin Medical University (Accession number: GLMC202103005). We consent for the publication of identifying images that compromise anonymity. Consent for publication: Not Applicable. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
The mitochondrial genome of Paragyrodactylus variegatus (Platyhelminthes: Monogenea): differences in major non-coding region and gene order compared to Gyrodactylus.Parasit Vectors. 2014 Aug 17;7:377. doi: 10.1186/1756-3305-7-377. Parasit Vectors. 2014. PMID: 25130627 Free PMC article.
-
Supplemental description of Gyrodactylus pseudorasborae (Gyrodactylidae) parasitic on topmouth gudgeon Pseudorasbora parva (Cyprinidae) in South China.Parasitol Int. 2024 Feb;98:102817. doi: 10.1016/j.parint.2023.102817. Epub 2023 Oct 16. Parasitol Int. 2024. PMID: 37852573
-
The first next-generation sequencing approach to the mitochondrial phylogeny of African monogenean parasites (Platyhelminthes: Gyrodactylidae and Dactylogyridae).BMC Genomics. 2018 Jul 4;19(1):520. doi: 10.1186/s12864-018-4893-5. BMC Genomics. 2018. PMID: 29973152 Free PMC article.
-
Sequencing of the complete mitochondrial genome of a fish-parasitic flatworm Paratetraonchoides inermis (Platyhelminthes: Monogenea): tRNA gene arrangement reshuffling and implications for phylogeny.Parasit Vectors. 2017 Oct 10;10(1):462. doi: 10.1186/s13071-017-2404-1. Parasit Vectors. 2017. PMID: 29017532 Free PMC article.
-
The complete mitochondrial genome of Gyrodactylus gurleyi (Platyhelminthes: Monogenea).Mitochondrial DNA B Resour. 2016 Jun 20;1(1):383-385. doi: 10.1080/23802359.2016.1172042. Mitochondrial DNA B Resour. 2016. PMID: 33490396 Free PMC article.
References
-
- Chen YY. Fuana Sinica Osteichthyes Cypriniformes II. Beijing, China: Science; 1998. (In Chinese).
-
- Onikura N, Nakajima J. Age, growth and habitat use of the topmouth gudgeon, Pseudorasbora parva in irrigation ditches on northwestern Kyushu Island, Japan. J Appl Ichthyol. 2013;29:186–92. 10.1111/j.1439-0426.2012.02041.x.
-
- Zheng WJ, Qin CB, Hu JY, Zhu YN, Yan X, Yang F, Lu RH, Yang LP. Morphological difference of Pseudorasbora parva in Yunnan, Irrawaddy River. J Anhui Agricultural Univ. 2017;44:248–53. 10.13610/j.cnki.1672-352x.20170419.025. (In Chinese).
-
- Jia YT, Kennard MJ, Liu YH, Sui XY, Chen YY, Li KM, Wang GJ, Chen YF. Understanding invasion success of Pseudorasbora parva in the Qinghai-Tibetan Plateau: insights from life-history and environmental filters. Sci Total Environ. 2019;694:133739. 10.1016/j.scitotenv.2019.133739. - PubMed
MeSH terms
Substances
Grants and funding
- 202310601023/College Students' Innovative Entrepreneurial Training Plan Program
- 20502018009/Basic Ability Enhancement Program for Young and Middle-aged Teachers of Guilin Medical University
- 31872203/Natural Science Foundation of China
- Guikeneng 22-A-02-01/Guangxi Key Laboratory of Rare and Endangered Animal Ecology, Guangxi Normal University
- 2023KY0524/Basic Ability Enhancement Program for Young and Middle-aged Teachers of Guangxi
LinkOut - more resources
Full Text Sources
Miscellaneous

