Genome-wide association study identifies candidate genes affecting body conformation traits of Zhongwei goat
- PMID: 39810085
- PMCID: PMC11730152
- DOI: 10.1186/s12864-024-11097-1
Genome-wide association study identifies candidate genes affecting body conformation traits of Zhongwei goat
Abstract
Background: Identifying markers or genes crucial for growth traits in Zhongwei goats is pivotal for breeding. Pinpointing genetic factors linked to body size gain enhances breeding efficiency and economic value. In this study, we used the MGISEQ-T7 platform to re-sequence 240 Zhongwei goats (133 male; 107 female) belonging to 5 metrics of growth traits at different growth stages (40 days and 6 months, here in after referred to as 40d and 6 m), namely, Body Weight (BW), Body Height (BH), Body Length (BL), Chest Circumference (CC), Tube Circumference (TC) were examined. A Genome-wide association study (GWAS) was conducted to identify candidate genes associated with the five indicators of body conformation traits, thereby establishing a foundation for subsequent investigations into the biological functions of these genes.
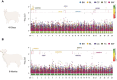
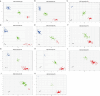
Results: A total of 19.89 Tb of raw data was generated with an average sequencing depth of about 20×. After quality control, 15,958,716 SNPs were available for the analysis. A total of 342 genome-wide significant SNPs were obtained. Among them, in the two physiological stages of 40d and 6 m, 147 and 32 SNPs were significantly associated with BW; 1 and 4 SNPs were significantly associated with BH; 19 and 6 SNPs were significantly associated with BL; 33 and 64 SNPs were significantly associated with CC, 34 and 2 SNPs were significantly associated with TC. These SNPs were annotated to 425 candidate genes. Finally, A total of 39 candidate genes are closely related to biological processes such as skeletal muscle development, skeletal formation, carcass quality, and embryonic development, where ADIPOQ, CCDD39, PTPRT, ZNF215, VRTN, ABCD4, DLST, ADAMTS2, ROBO1, AKAP13, AQPI, SOX2, and AHSG were identified as an important component of the genetic framework that may control somatic conformational traits in Zhongwei goats. which warrants further investigation and review. We verified the polymorphism of 11 SNPs by KASP, and found that Chr13_g.11,700,438 A > G, Chr15_g.37,120,328 A > G, Chr6_g.7,209,383 C > T, Chr20_g.51277932T > A, Chr19_g.17,078,199 A > G, and Chr1_g.79,943,276 C > T were significantly genotyped in verified populations (P < 0.001).
Conclution: It is the first GWAS study to analyze genomic data from 40d and 6 m of Zhongwei goats to understand the molecular genetic mechanisms of growth. Our research identified a series of SNPs and candidate genes associated with growth traits, which could assist us in developing the meat production trait in Zhongwei goats.
Keywords: Body conformation traits; GWAS; KASP validation; Re-sequence; Zhongwei goat.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All samples were collected in accordance with the International Guiding Principles for Biomedical Research involving animals and approved by the Special Committee on Scientific Research and Academic Ethics of Inner Mongolia Agricultural University, responsible for the approval of Biomedical Research Ethics of Inner Mongolia Agricultural University [Approval No. (2020) 056]. No specific permissions were required for these activities, and no endangered or protected species were involved. These activities were performed under the informed consent from all owners. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Genome-wide association study for cashmere traits in Inner Mongolia cashmere goat population reveals new candidate genes and haplotypes.BMC Genomics. 2024 Jul 2;25(1):658. doi: 10.1186/s12864-024-10543-4. BMC Genomics. 2024. PMID: 38956486 Free PMC article.
-
Genome-wide association study of growth and reproductive traits based on low-coverage whole-genome sequencing in a Chubao black-head goat population.Gene. 2024 Dec 30;931:148891. doi: 10.1016/j.gene.2024.148891. Epub 2024 Aug 24. Gene. 2024. PMID: 39187139
-
Identification of Genomic Regions and Candidate Genes Associated with Body Weight and Body Conformation Traits in Karachai Goats.Genes (Basel). 2022 Sep 30;13(10):1773. doi: 10.3390/genes13101773. Genes (Basel). 2022. PMID: 36292658 Free PMC article.
-
Whole genome and transcriptome analyses identify genetic markers associated with growth traits in Qinchuan black pig.BMC Genomics. 2025 May 12;26(1):469. doi: 10.1186/s12864-025-11627-5. BMC Genomics. 2025. PMID: 40355827 Free PMC article.
-
Potential Candidate Genes Associated with Litter Size in Goats: A Review.Animals (Basel). 2025 Jan 2;15(1):82. doi: 10.3390/ani15010082. Animals (Basel). 2025. PMID: 39795025 Free PMC article. Review.
Cited by
-
Genome-Wide Association Study for Weight-Related Traits in Scylla paramamosain Using Whole-Genome Resequencing.Animals (Basel). 2025 Jun 20;15(13):1829. doi: 10.3390/ani15131829. Animals (Basel). 2025. PMID: 40646730 Free PMC article.
-
Rapid Identification of the SNP Mutation in the ABCD4 Gene and Its Association with Multi-Vertebrae Phenotypes in Ujimqin Sheep Using TaqMan-MGB Technology.Animals (Basel). 2025 Aug 5;15(15):2284. doi: 10.3390/ani15152284. Animals (Basel). 2025. PMID: 40805073 Free PMC article.
References
MeSH terms
Grants and funding
- 2023AAC03898/Ningxia Natural Science Foundation
- 2021YFD1200902/National Key Research and Development Plan Project Sub-project
- NMGIRT2322/Supported by Program for Innovative Research Team in Universities of Inner Mongolia Autonomous Region
- BR22-13-02/Basic scientific research business fee project for universities directly under the Inner Mongolia Autonomous Region
LinkOut - more resources
Full Text Sources
Miscellaneous

