Genome-wide identification and functional roles relating to anthocyanin biosynthesis analysis in maize
- PMID: 39810086
- PMCID: PMC11734362
- DOI: 10.1186/s12870-025-06053-4
Genome-wide identification and functional roles relating to anthocyanin biosynthesis analysis in maize
Abstract
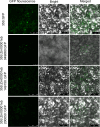
Background: Anthocyanin is an important class of water-soluble pigments that are widely distributed in various tissues of plants, and it not only facilitates diverse color changes but also plays important roles in various biological processes. Maize silk, serving as an important reproductive organ and displaying a diverse range of colors, plays an indispensable role in biotic resistance through its possession of anthocyanin. However, the copy numbers, characteristics, and expression patterns of genes involved in maize anthocyanin biosynthesis are not fully understood. In this study, gene numbers, distribution, structure, cis-elements of the anthocyanin synthetic gene family were identified, and then the potential transcriptional factors were predicted by two analyzed methods. Finally, genes involved in maize silk pigment were screened by un-targeted metabolism analysis.
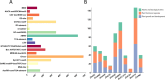
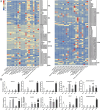
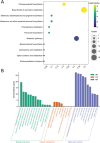
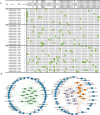
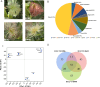
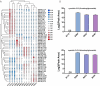
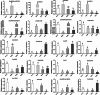
Results: Ten gene families involved in the maize anthocyanin biosynthesis pathway were identified, and 142 synthetic genes were obtained. These anthocyanin biosynthetic genes have high copy numbers and are normally clustered on chromosomes. The promoters of these synthetic genes contain various cis-elements and the gene expression patterns and transcriptional regulatory networks were analyzed. These genes are distributed on different chromosomes and gene expression patterns vary across different tissues in maize. Specifically, these genes often exhibit higher expression in the stem, leaves, and seeds. Ten highly expressed genes in silks were identified. Based on un-targeted metabolites detection in the silks of four maize representative inbred lines with different colors, two main differential anthocyanin components were identified. Furthermore, the gene expression patterns of the ten highly expressed genes and their potential interacting transcriptional factors were analyzed across the four inbred lines.
Conclusions: The results in this study show a through picture of maize anthocyanin synthetic genes, and the structure and function of genes related to anthocyanin biosynthesis in maize could be further investigated.
Keywords: Anthocyanin; Biosynthetic genes; Gene expression; Maize; Silk.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests. Clinical trial number: Not applicable.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

