TP53 germline testing and hereditary cancer: how somatic events and clinical criteria affect variant detection rate
- PMID: 39810221
- PMCID: PMC11734529
- DOI: 10.1186/s13073-025-01429-5
TP53 germline testing and hereditary cancer: how somatic events and clinical criteria affect variant detection rate
Abstract
Background: Germline heterozygous pathogenic variants (PVs) in TP53 cause Li-Fraumeni syndrome (LFS), a condition associated with increased risk of multiple tumor types. As the associated cancer risks were refined over time, clinical criteria also evolved to optimize diagnostic yield. The implementation of multi-gene panel germline testing in different clinical settings has led to the identification of TP53 PV carriers outside the classic LFS-associated cancer phenotypes, leading to a broader cancer phenotypic redefinition and to the renaming of the condition as "heritable TP53-related cancer syndrome" (hTP53rc). Germline TP53 variant interpretation is challenging due to the diverse nature of TP53 PVs, variable penetrance of the syndrome, possible occurrence of TP53 somatic mosaicism, and TP53 involvement in clonal hematopoiesis of indeterminate potential (CHIP). Here we aim to assess the relevance and impact of these issues on the diagnostic routine, and to evaluate the sensitivity of the different LFS clinical criteria to identify hTP53rc.
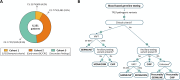
Methods: TP53 was analyzed in 6161 suspected hereditary cancer non-related patients categorized into three subgroups: (1) 495 patients fulfilling any LFS/Chompret clinical criteria; (2) 2481 patients diagnosed with early-onset breast/colorectal cancer; (3) 3185 patients without clinical criteria suggestive of hTP53rc. Ancillary tests were performed when TP53 PVs were identified in individuals not meeting LFS/Chompret criteria and/or when the variant was identified at low variant allele frequency (VAF).
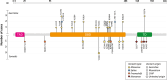
Results: TP53 PVs were identified in blood DNA of 45 probands. Variant origin was elucidated in 39 of these: 72% patients had a constitutional PV, 10% were mosaics, and 18% had CHIP-associated PVs. Notably, two of the seven CHIP-TP53 PVs identified were detected at high allelic frequencies (VAF > 35%). Twenty-nine percent of germline TP53 PV did not meet any of the LFS clinical criteria. Among the clinical criteria, Chompret 2009 showed the highest sensitivity in our cohort (68% vs. 54% for Chompret 2015), highlighting the relevance of considering lung cancer in the criteria.
Conclusions: Our data supports performing TP53 ancillary testing for the identification of potential mosaicisms and CHIP-associated PVs, particularly in patients not meeting clinical criterial for LFS, irrespective of the VAF, and the application of clinical criteria that include lung cancer diagnosis.
Keywords: TP53; Clonal hematopoiesis; Hereditary cancer; Heritable TP53-related cancer syndrome; Li-Fraumeni syndrome; Mosaicism.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The research was conducted in accordance with the principles of the Declaration of Helsinki, and ethical approval was obtained from the ethics committee of Bellvitge Biomedical Research Institute (IDIBELL; PR278/19). Informed written consent for both diagnostic and research purposes was obtained from all participants. Consent for publication: Written informed consent for publication was obtained from all study participants. Competing interests: The authors declare no competing interests.
Figures


References
-
- Muller PAJ, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25(3):304–17. Available from: https://pubmed.ncbi.nlm.nih.gov/24651012/. Cited 2024 Jul 18. - PMC - PubMed
-
- Mai PL, Best AF, Peters JA, DeCastro RM, Khincha PP, Loud JT, et al. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer. 2016;122(23):3673–81. Available from: https://pubmed.ncbi.nlm.nih.gov/27496084/. Cited 2024 Feb 5. - PMC - PubMed
-
- Gonzalez KD, Noltner KA, Buzin CH, Gu D, Wen-Fong CY, Nguyen VQ, et al. Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol. 2009;27(8):1250–6. Available from: https://pubmed.ncbi.nlm.nih.gov/19204208/. Cited 2024 Jun 18. - PubMed
-
- Chompret A, Abel A, Stoppa-Lyonnet D, Brugières L, Pagès S, Feunteun J, et al. Sensitivity and predictive value of criteria for p53 germline mutation screening. J Med Genet. 2001;38(1):43–7. Available from: https://pubmed.ncbi.nlm.nih.gov/11332399/. Cited 2024 Feb 5. - PMC - PubMed
-
- Tinat J, Bougeard G, Baert-Desurmont S, Vasseur S, Martin C, Bouvignies E, et al. 2009 version of the Chompret criteria for Li Fraumeni syndrome. J Clin Oncol. 2009;27(26). Available from: https://pubmed.ncbi.nlm.nih.gov/19652052/. Cited 2024 Feb 5. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

