Enhancing NK cell-mediated tumor killing of B7-H6+ cells with bispecific antibodies targeting allosteric sites of NKp30
- PMID: 39811682
- PMCID: PMC11730255
- DOI: 10.1016/j.omton.2024.200917
Enhancing NK cell-mediated tumor killing of B7-H6+ cells with bispecific antibodies targeting allosteric sites of NKp30
Abstract
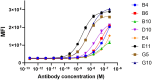
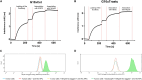
In this work, we report the discovery and engineering of allosteric variable domains of the heavy chain (VHHs) derived from camelid immunization targeting NKp30, an activating receptor on natural killer (NK) cells. The aim was to enhance NK cell-mediated killing capacities by identifying VHHs that do not compete with the natural ligand of NKp30:B7-H6, thereby maximizing the recognition of B7-H6+ tumor cells. By relying on the DuoBody technology, bispecific therapeutic antibodies were engineered, creating a panel of bispecific antibodies against NKp30xEGFR (cetuximab moiety) or NKp30xHER2 (trastuzumab moiety), called natural killer cell engagers (NKCEs). These NKCEs were assessed for their killing capacities on B7-H6-expressing tumor cells. The results demonstrated an enhancement in NK killing capacities for both EGFR-expressing (HeLa) and HER2-expressing (SK-BR-3) cells, indicating the significance of the natural NKp30/B7-H6 axis in tumor recognition by the immune system. Notably, engineering NKCEs to allow natural recognition of B7-H6 was found to be more effective in promoting NKCE-mediated killing of B7-H6+ tumor cells via enhancement of cytokine release. This study highlights the potential of an enhanced-targeting approach, wherein tumor cell surface antigens are targeted while still enabling the natural recognition of the activating ligand (B7-H6) by the immune cells.
Keywords: DuoBody; EGFR; MT: Regular Issue; NKCE; NKp30; allosteric site; bispecific antibodies; bsAbs; cancer therapy; natural-killer cell engager.
© 2024 The Author(s).
Conflict of interest statement
L.F., P.A., L.P., S.Z., L.T., and S.B. are employees of Merck Healthcare KGaA.
Figures




References
-
- Rebuffet L., Melsen J.E., Escalière B., Basurto-Lozada D., Bhandoola A., Björkström N.K., Bryceson Y.T., Castriconi R., Cichocki F., Colonna M., et al. High-dimensional single-cell analysis of human natural killer cell heterogeneity. Nat. Immunol. 2024;25:1474–1488. doi: 10.1038/s41590-024-01883-0. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
