Host-microbe multi-omics and succinotype profiling have prognostic value for future relapse in patients with inflammatory bowel disease
- PMID: 39812341
- PMCID: PMC11740686
- DOI: 10.1080/19490976.2025.2450207
Host-microbe multi-omics and succinotype profiling have prognostic value for future relapse in patients with inflammatory bowel disease
Abstract
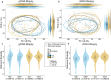
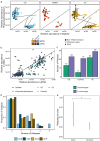
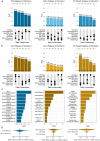
Crohn's disease (CD) and ulcerative colitis (UC) are chronic relapsing inflammatory bowel disorders (IBD), the pathogenesis of which is uncertain but includes genetic susceptibility factors, immune-mediated tissue injury and environmental influences, most of which appear to act via the gut microbiome. We hypothesized that host-microbe alterations could be used to prognostically stratify patients experiencing relapses up to four years after endoscopy. We therefore examined multiple omics data, including published and new datasets, generated from paired inflamed and non-inflamed mucosal biopsies from 142 patients with IBD (54 CD; 88 UC) and from 34 control (non-diseased) biopsies. The relapse-predictive potential of 16S rRNA gene and transcript amplicons (standing and active microbiota) were investigated along with host transcriptomics, epigenomics and genetics. While standard single-omics analysis could not distinguish between patients who relapsed and those that remained in remission within four years of colonoscopy, we did find an association between the number of flares and a patient's succinotype. Our multi-omics machine learning approach was also able to predict relapse when combining features from the microbiome and human host. Therefore multi-omics, rather than single omics, better predicts relapse within 4 years of colonoscopy, while a patient's succinotype is associated with a higher frequency of relapses.
Keywords: Crohn’s disease; gut microbiome; host-microbe interactions; inflammatory bowel disease; machine learning; ulcerative colitis.
Conflict of interest statement
G.E.L is an employee and shareholder of PharmaBiome AG and inventor on the patent application WO 2023/118460 A1 entitled “New biomarker for disorders and diseases associated with intestinal dysbiosis”. M.J.C. is co-founder and Head of Bioinformatics of SeqBiome. T.Z.D. is a co-founder and was Vice-president of Second Genome; S.I., K.D., and E.R. were employees of Second Genome at the time of the analysis. F.S. is a co-founder of three campus companies: Alimentary Health Ltd, Tuscan Health Ltd (now names 4D Pharma Cork) and Alantia Food Clinical Trials.
Figures



References
-
- Clooney AG, Eckenberger J, Laserna-Mendieta E, Sexton KA, Bernstein MT, Vagianos K, Sargent M, Ryan FJ, Moran C, Sheehan D, et al. Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut. 2021;70(3):499–510. doi:10.1136/gutjnl-2020-321106. - DOI - PMC - PubMed
-
- Ryan FJ, Ahern AM, Fitzgerald RS, Laserna-Mendieta EJ, Power EM, Clooney AG, O’Donoghue KW, McMurdie PJ, Iwai S, Crits-Christoph A, et al. Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat Commun. 2020;11(1):1–12. doi:10.1038/s41467-020-15342-5. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
