Nucleosome repositioning in cardiac reprogramming
- PMID: 39813277
- PMCID: PMC11734986
- DOI: 10.1371/journal.pone.0317718
Nucleosome repositioning in cardiac reprogramming
Abstract
Early events in the reprogramming of fibroblasts to cardiac muscle cells are unclear. While various histone undergo modification and re-positioning, and these correlate with the activity of certain genes, it is unknown if these events are causal or happen in response to reprogramming. Histone modification and re-positioning would be expected to open up chromatin on lineage-specific genes and this can be ascertained by studying nucleosome architecture. We have recently developed a set of tools to identify significant changes in nucleosome architecture which we used to study skeletal muscle differentiation. In this report, we have applied these tools to understand nucleosome architectural changes during fibroblast to cardiac muscle reprogramming. We found that nucleosomes surrounding the transcription start sites of cardiac muscle genes induced during reprogramming were insensitive to reprogramming factors as well as to agents which enhance reprogramming efficacy. In contrast, significant changes in nucleosome architecture were observed distal to the transcription start site. These regions were associated with nucleosome build-up. In summary, investigations into nucleosome structure do not support the notion that fibroblasts to cardiac muscle cell reprogramming involves chromatin opening and suggests instead long-range effects such as breaking closed-loop inhibition.
Copyright: © 2025 Harris et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

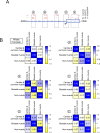
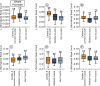
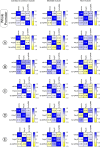

Update of
-
Nucleosome repositioning in cardiac reprogramming.bioRxiv [Preprint]. 2024 Nov 6:2024.11.05.622077. doi: 10.1101/2024.11.05.622077. bioRxiv. 2024. Update in: PLoS One. 2025 Jan 15;20(1):e0317718. doi: 10.1371/journal.pone.0317718. PMID: 39574721 Free PMC article. Updated. Preprint.
References
-
- Jayawardena TM, Finch EA, Zhang L, Zhang H, Hodgkinson CP, Pratt RE, et al.. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ Res. 2015;116(3):418–24. Epub 2014/10/30. doi: 10.1161/CIRCRESAHA.116.304510 ; PubMed Central PMCID: PMC4312531. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

