SMARCA4 regulates the NK-mediated killing of senescent cells
- PMID: 39813356
- PMCID: PMC11734740
- DOI: 10.1126/sciadv.adn2811
SMARCA4 regulates the NK-mediated killing of senescent cells
Abstract
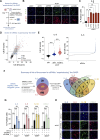
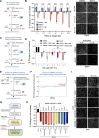
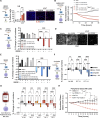
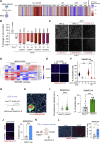
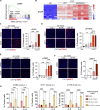
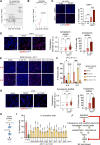
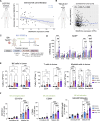
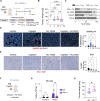
Induction of senescence by chemotherapeutic agents arrests cancer cells and activates immune surveillance responses to contribute to therapy outcomes. In this investigation, we searched for ways to enhance the NK-mediated elimination of senescent cells. We used a staggered screen approach, first identifying siRNAs potentiating the secretion of immunomodulatory cytokines to later test for their ability to enhance NK-mediated killing of senescent cells. We identified that genetic or pharmacological inhibition of SMARCA4 enhanced senescent cell elimination by NK cells. SMARCA4 expression is elevated during senescence and its inhibition derepresses repetitive elements, inducing the SASP via activation of cGAS/STING and MAVS/MDA5 pathways. Moreover, a PROTAC targeting SMARCA4 synergized with cisplatin to increase the infiltration of CD8 T cells and mature, activated NK cells in an immunocompetent model of ovarian cancer. Our results indicate that SMARCA4 inhibitors enhance NK-mediated surveillance of senescent cells and may represent senotherapeutic interventions for ovarian cancer.
Figures
References
-
- Gorgoulis V., Adams P. D., Alimonti A., Bennett D. C., Bischof O., Bishop C., Campisi J., Collado M., Evangelou K., Ferbeyre G., Gil J., Hara E., Krizhanovsky V., Jurk D., Maier A. B., Narita M., Niedernhofer L., Passos J. F., Robbins P. D., Schmitt C. A., Sedivy J., Vougas K., von Zglinicki T., Zhou D., Serrano M., Demaria M., Cellular senescence: Defining a path forward. Cell 179, 813–827 (2019). - PubMed
-
- Faget D. V., Ren Q., Stewart S. A., Unmasking senescence: Context-dependent effects of SASP in cancer. Nat. Rev. Cancer 19, 439–453 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

