Progressive natural killer cell dysfunction in advanced-stage clear-cell renal cell carcinoma and association with clinical outcomes
- PMID: 39813824
- PMCID: PMC11783098
- DOI: 10.1016/j.esmoop.2024.104105
Progressive natural killer cell dysfunction in advanced-stage clear-cell renal cell carcinoma and association with clinical outcomes
Abstract
Background: Natural killer (NK) cells are important contributors to antitumor immunity in clear-cell renal cell carcinoma (ccRCC). However, their phenotype, function, and association with clinical outcomes in ccRCC remain poorly understood.
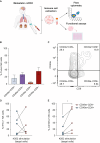
Materials and methods: We analyzed single-cell RNA sequencing data from 13 primary tumors, 1 localized tumor extension, and 1 metastasis from ccRCC patients at different clinical stages. For each primary tumor specimen, paired normal kidneys were also analyzed. Differential gene expression analysis was carried out to investigate NK cell phenotypes and to derive a gene expression signature. Gene signatures from NK cell subclusters of interest were used to interrogate bulk transcriptomic datasets and expression with clinical outcomes. Finally, tumor-infiltrating NK cell function (cytokine production and cytotoxicity) was assessed by isolation of live NK cells from ccRCC tissue, co-culture with K562 target cells, and measurement of cytokine production (interferon-γ) and cytotoxicity (CD107a) markers by flow cytometry.
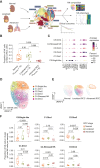
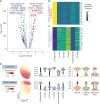
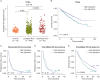
Results: Single-cell transcriptomic data were analyzed from 13 patients with ccRCC (tumor/normal kidney), resulting in 21 139 NK cells. Clustering analysis revealed six NK cell subsets. Bright-like NK cells were significantly enriched in advanced ccRCC compared with localized ccRCC and normal kidney, expressed markers of tissue residency (ZNF683/Hobit, ITGA1/CD49a, CD9, ITGAE/CD103), and had decreased expression of cytotoxicity genes (GZMB/Granzyme-B, PRF1/perforin). In independent cohorts (The Cancer Genome Atlas ccRCC cohort, CheckMate 025), a gene expression score representing this dysfunctional NK cell phenotype was enriched in advanced ccRCC and was associated with worse overall survival. Functional interrogation of tumor-infiltrating NK cells from ccRCC confirmed that tumor-resident CD49a+CD9+ NK cells had impaired cytotoxicity compared with CD49a-CD9- NK cells.
Conclusions: A dysfunctional, tumor-resident NK cell phenotype was enriched among patients with metastatic disease and associated with worse survival in patients with advanced ccRCC across multiple patient cohorts. Restoration of NK cell function (via cytokine stimulation or NK cell engineering) could provide a novel avenue for therapeutic intervention against ccRCC.
Keywords: NK cells; immunogenomics; renal cell carcinoma; single-cell transcriptomics.
Copyright © 2025. Published by Elsevier Ltd.
Figures
References
-
- Bakouny Z., Flippot R., Braun D.A., Lalani A.-K.A., Choueiri T.K. State of the future: translational approaches in renal cell carcinoma in the immunotherapy era. Eur Urol Focus. 2020;6:37–40. - PubMed
-
- Xu W., Atkins M.B., McDermott D.F. Checkpoint inhibitor immunotherapy in kidney cancer. Nat Rev Urol. 2020;17:137–150. - PubMed
-
- Choueiri T.K., Motzer R.J. Systemic therapy for metastatic renal-cell carcinoma. N Engl J Med. 2017;376:354–366. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

