UNC119 regulates T-cell receptor signalling in primary T cells and T acute lymphocytic leukaemia
- PMID: 39814552
- PMCID: PMC11735834
- DOI: 10.26508/lsa.202403066
UNC119 regulates T-cell receptor signalling in primary T cells and T acute lymphocytic leukaemia
Abstract
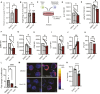
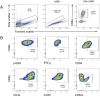
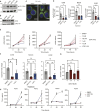
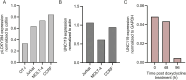
T-cell receptor recognition of cognate peptide-MHC leads to the formation of signalling domains and the immunological synapse. Because of the close membrane apposition, there is rapid exclusion of CD45, and therefore LCK activation. Much less is known about whether spatial regulation of the intracellular face dictates LCK activity and TCR signal transduction. Moreover, as LCK is a driver in T acute lymphocytic leukaemia, it is important to understand its regulation. Here, we demonstrate a direct role of the ciliary protein UNC119 in trafficking LCK to the immunological synapse. Inhibiting UNC119 reduces localisation of LCK without impairing LCK phosphorylation and reduces T-cell receptor signal transduction. Although important for initial LCK reorganisation, activated CD8+ T cells retained their ability to kill target tumour cells when UNC119 was inhibited. UNC119 was also needed to sustain proliferation in patient-derived T-ALL cells. UNC119 may therefore represent a novel therapeutic target in T acute lymphocytic leukaemia, which alters the subcellular localisation of LCK in T acute lymphocytic leukaemia cells but preserves the function of existing cytotoxic lymphocytes.
© 2025 Samarakoon et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
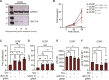
Figures





Similar articles
-
The Ciliary Machinery Is Repurposed for T Cell Immune Synapse Trafficking of LCK.Dev Cell. 2018 Oct 8;47(1):122-132.e4. doi: 10.1016/j.devcel.2018.08.012. Epub 2018 Sep 13. Dev Cell. 2018. PMID: 30220567 Free PMC article.
-
HIV-1 Nef compensates for disorganization of the immunological synapse by inducing trans-Golgi network-associated Lck signaling.Blood. 2012 Jan 19;119(3):786-97. doi: 10.1182/blood-2011-08-373209. Epub 2011 Nov 28. Blood. 2012. PMID: 22123847
-
Uncoordinated 119 protein controls trafficking of Lck via the Rab11 endosome and is critical for immunological synapse formation.J Immunol. 2009 Aug 1;183(3):1675-84. doi: 10.4049/jimmunol.0900792. Epub 2009 Jul 10. J Immunol. 2009. PMID: 19592652
-
Consequences of a mutation in the UNC119 gene for T cell function in idiopathic CD4 lymphopenia.Curr Allergy Asthma Rep. 2012 Oct;12(5):396-401. doi: 10.1007/s11882-012-0281-4. Curr Allergy Asthma Rep. 2012. PMID: 22729960 Review.
-
Lipid rafts in T cell receptor signalling.Mol Membr Biol. 2006 Jan-Feb;23(1):49-57. doi: 10.1080/09687860500453673. Mol Membr Biol. 2006. PMID: 16611580 Free PMC article. Review.
References
-
- Williams BL, Schreiber KL, Zhang W, Wange RL, Samelson LE, Leibson PJ, Abraham RT (1998) Genetic evidence for differential coupling of syk family kinases to the T-cell receptor: Reconstitution studies in a ZAP-70-deficient Jurkat T-cell line. Mol Cell Biol 18: 1388–1399. 10.1128/MCB.18.3.1388 - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
