Microglial NLRP3-gasdermin D activation impairs blood-brain barrier integrity through interleukin-1β-independent neutrophil chemotaxis upon peripheral inflammation in mice
- PMID: 39814731
- PMCID: PMC11735931
- DOI: 10.1038/s41467-025-56097-1
Microglial NLRP3-gasdermin D activation impairs blood-brain barrier integrity through interleukin-1β-independent neutrophil chemotaxis upon peripheral inflammation in mice
Abstract
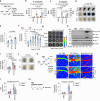
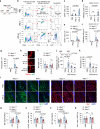
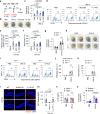
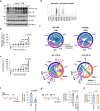
Blood-brain barrier (BBB) disintegration is a key contributor to neuroinflammation; however, the biological processes governing BBB permeability under physiological conditions remain unclear. Here, we investigate the role of NLRP3 inflammasome in BBB disruption following peripheral inflammatory challenges. Repeated intraperitoneal lipopolysaccharide administration causes NLRP3-dependent BBB permeabilization and myeloid cell infiltration into the brain. Using a mouse model with cell-specific hyperactivation of NLRP3, we identify microglial NLRP3 activation as essential for peripheral inflammation-induced BBB disruption. Conversely, NLRP3 and microglial gasdermin D (GSDMD) deficiency markedly attenuates lipopolysaccharide-induced BBB breakdown. Notably, IL-1β is not required for NLRP3-GSDMD-mediated BBB disruption. Instead, microglial NLRP3-GSDMD axis upregulates CXCL chemokines and matrix metalloproteinases around BBB via producing GDF-15, promoting the recruitment of CXCR2-containing neutrophils. Inhibition of neutrophil infiltration and matrix metalloproteinase activity significantly reduces NLRP3-mediated BBB impairment. Collectively, these findings reveal the important role of NLRP3-driven chemokine production in BBB disintegration, suggesting potential therapeutic targets to mitigate neuroinflammation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Abbott, N. J., Ronnback, L. & Hansson, E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci.7, 41–53 (2006). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

