A preliminary evaluation of a fast, low-cost, and high-throughput nucleic acid extraction method for bacterial microbiota profiling in low-microbial biomass samples
- PMID: 39814772
- PMCID: PMC11736122
- DOI: 10.1038/s41598-024-84682-9
A preliminary evaluation of a fast, low-cost, and high-throughput nucleic acid extraction method for bacterial microbiota profiling in low-microbial biomass samples
Abstract
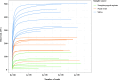
The respiratory tract is colonized with low-density microbial communities, which have been shown to impact human respiratory health through microbiota-host interactions. However, a lack of fast and cost-effective nucleic acid extraction method for low-microbial biomass samples hinders investigation of respiratory microbiota. Here, we performed a pilot study to assess the suitability of the NAxtra nucleic acid extraction protocol for profiling bacterial microbiota in respiratory samples. A small number of nasopharyngeal aspirate (n = 8), nasal swab (n = 8), and saliva samples (n = 8) were collected, nucleic acids were isolated using the NAxtra protocol, and 16 S rRNA gene sequencing was performed to characterize bacterial microbiota, which were compared to the same sample types from previous studies using other protocols. The bacterial composition in nasal and saliva samples were consistent with previous reports. Saliva microbiota was significantly richer than nasal microbiota and varied less among individual samples than nasal microbiota. Bacterial composition in nasal samples was distinct from nasopharyngeal aspirates, but closer to saliva samples. A sequencing depth of 50,000 reads/sample was sufficient for microbiota profiling in low biomass respiratory samples. Our pilot study indicates the potential of the NAxtra protocol for bacterial microbiota characterization of low-microbial biomass samples and supports a more comprehensive study to fully evaluate the value of the NAxtra protocol in microbiota research and clinical diagnostics of respiratory pathogens.
Keywords: 16S rRNA gene sequencing; Bacteria; Low-microbial biomass; Microbiota; Nucleic acid extraction; Respiratory samples.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

