Specification of claustro-amygdalar and palaeocortical neurons and circuits
- PMID: 39814878
- PMCID: PMC11821539
- DOI: 10.1038/s41586-024-08361-5
Specification of claustro-amygdalar and palaeocortical neurons and circuits
Abstract
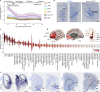
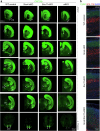
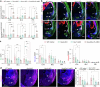
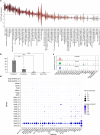
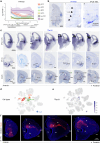
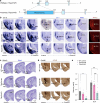
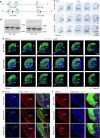
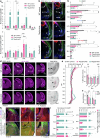
The ventrolateral pallial (VLp) excitatory neurons in the claustro-amygdalar complex and piriform cortex (PIR; which forms part of the palaeocortex) form reciprocal connections with the prefrontal cortex (PFC), integrating cognitive and sensory information that results in adaptive behaviours1-5. Early-life disruptions in these circuits are linked to neuropsychiatric disorders4-8, highlighting the importance of understanding their development. Here we reveal that the transcription factors SOX4, SOX11 and TFAP2D have a pivotal role in the development, identity and PFC connectivity of these excitatory neurons. The absence of SOX4 and SOX11 in post-mitotic excitatory neurons results in a marked reduction in the size of the basolateral amygdala complex (BLC), claustrum (CLA) and PIR. These transcription factors control BLC formation through direct regulation of Tfap2d expression. Cross-species analyses, including in humans, identified conserved Tfap2d expression in developing excitatory neurons of BLC, CLA, PIR and the associated transitional areas of the frontal, insular and temporal cortex. Although the loss and haploinsufficiency of Tfap2d yield similar alterations in learned threat-response behaviours, differences emerge in the phenotypes at different Tfap2d dosages, particularly in terms of changes observed in BLC size and BLC-PFC connectivity. This underscores the importance of Tfap2d dosage in orchestrating developmental shifts in BLC-PFC connectivity and behavioural modifications that resemble symptoms of neuropsychiatric disorders. Together, these findings reveal key elements of a conserved gene regulatory network that shapes the development and function of crucial VLp excitatory neurons and their PFC connectivity and offer insights into their evolution and alterations in neuropsychiatric disorders.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures








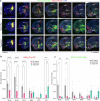
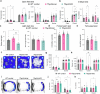

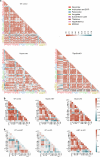
References
-
- Swanson, L. W. & Petrovich, G. D. What is the amygdala? Trends Neurosci.21, 323–331 (1998). - PubMed
-
- Amaral, D. G., Schumann, C. M. & Nordahl, C. W. Neuroanatomy of autism. Trends Neurosci.31, 137–145 (2008). - PubMed
-
- McDonald, A. J. Cortical pathways to the mammalian amygdala. Prog. Neurobiol.55, 257–332 (1998). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

