Autoactive CNGC15 enhances root endosymbiosis in legume and wheat
- PMID: 39814887
- PMCID: PMC11839481
- DOI: 10.1038/s41586-024-08424-7
Autoactive CNGC15 enhances root endosymbiosis in legume and wheat
Abstract
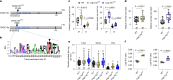
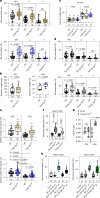
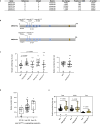
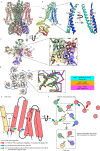
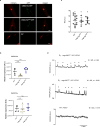
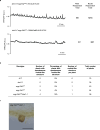
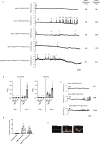
Nutrient acquisition is crucial for sustaining life. Plants develop beneficial intracellular partnerships with arbuscular mycorrhiza (AM) and nitrogen-fixing bacteria to surmount the scarcity of soil nutrients and tap into atmospheric dinitrogen, respectively1,2. Initiation of these root endosymbioses requires symbiont-induced oscillations in nuclear calcium (Ca2+) concentrations in root cells3. How the nuclear-localized ion channels, cyclic nucleotide-gated channel (CNGC) 15 and DOESN'T MAKE INFECTIONS1 (DMI1)4 are coordinated to specify symbiotic-induced nuclear Ca2+ oscillations remains unknown. Here we discovered an autoactive CNGC15 mutant that generates spontaneous low-frequency Ca2+ oscillations. While CNGC15 produces nuclear Ca2+ oscillations via a gating mechanism involving its helix 1, DMI1 acts as a pacemaker to specify the frequency of the oscillations. We demonstrate that the specificity of symbiotic-induced nuclear Ca2+ oscillations is encoded in its frequency. A high frequency activates endosymbiosis programmes, whereas a low frequency modulates phenylpropanoid pathways. Consequently, the autoactive cngc15 mutant, which is capable of generating both frequencies, has increased flavonoids that enhance AM, root nodule symbiosis and nutrient acquisition. We transferred this trait to wheat, resulting in field-grown wheat with increased AM colonization and nutrient acquisition. Our findings reveal a new strategy to boost endosymbiosis in the field and reduce inorganic fertilizer use while sustaining plant growth.
© 2025. Crown.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







Comment in
-
Calcium beats for better crops.Nat Plants. 2025 Feb;11(2):148. doi: 10.1038/s41477-025-01935-9. Nat Plants. 2025. PMID: 39948262 No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

