Loss of correlated proteasomal subunit expression selectively promotes the 20SHigh state which underlies luminal breast tumorigenicity
- PMID: 39814910
- PMCID: PMC11735796
- DOI: 10.1038/s42003-024-07432-7
Loss of correlated proteasomal subunit expression selectively promotes the 20SHigh state which underlies luminal breast tumorigenicity
Abstract
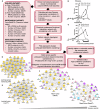
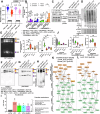
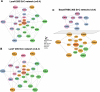
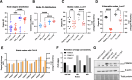
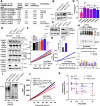
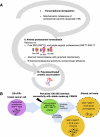
Why cancer cells disproportionately accumulate polyubiquitinated proteotoxic proteins despite high proteasomal activity is an outstanding question. While mis-regulated ubiquitination is a contributing factor, here we show that a structurally-perturbed and sub-optimally functioning proteasome is at the core of altered proteostasis in tumors. By integrating the gene coexpression signatures of proteasomal subunits in breast cancer (BrCa) patient tissues with the atomistic details of 26S holocomplex, we find that the transcriptional deregulation induced-stoichiometric imbalances perpetuate with disease severity. As seen in luminal BrCa cell lines, this imbalance limits the number of double-capped 19S-20S-19S holocomplexes (30S) formed and promotes free 20S catalytic core accumulation that is widely-believed to confer survival advantage to tumors. By retaining connectivity with key tumor 19S:20S interface nodes, the PSMD9 19S subunit chaperone emerges as a crucial regulator of 26S/30S:20S ratios sustaining tumor cell proteasome function. Disrupting this connectivity by depleting PSMD9 in MCF7 cells introduces structural anomalies in the proteasome, and shifts dependence from 20SHigh to a deregulated 26SHigh state invoking anti-tumor responses which opens up clinically-relevant therapeutic possibilities.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

