Multi-omic analysis of chronic myelomonocytic leukemia monocytes reveals metabolic and immune dysregulation leading to altered macrophage polarization
- PMID: 39815051
- PMCID: PMC11879862
- DOI: 10.1038/s41375-024-02511-4
Multi-omic analysis of chronic myelomonocytic leukemia monocytes reveals metabolic and immune dysregulation leading to altered macrophage polarization
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: PBMNC samples were obtained from CMML patients treated at The Christie NHS Foundation Trust (Manchester, UK) recruited to the Manchester Cancer Research Centre Tissue Biobank (initiated with the approval of South Manchester Research Ethics Committee). PBMNCs from age-matched Healthy volunteers were used as controls. All patients and controls gave informed consent according to the Declaration of Helsinki. The study was approved by the UK Health Research Authority (REC: 19/LO/0564).
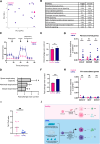
Figures

References
-
- Itzykson R, Kosmider O, Renneville A, Morabito M, Preudhomme C, Berthon C, et al. Clonal architecture of chronic myelomonocytic leukemias. Blood. 2013;121:2186–98. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

