Single-cell microbiota phenotyping reveals distinct disease and therapy-associated signatures in Crohn's disease
- PMID: 39815413
- PMCID: PMC11740678
- DOI: 10.1080/19490976.2025.2452250
Single-cell microbiota phenotyping reveals distinct disease and therapy-associated signatures in Crohn's disease
Abstract
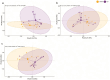
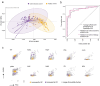
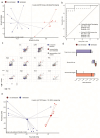
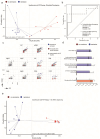
IgA-coated fractions of the intestinal microbiota of Crohn's disease (CD) patients have been shown to contain taxa that hallmark the compositional dysbiosis in CD microbiomes. However, the correlation between other cellular properties of intestinal bacteria and disease has not been explored further, especially for features that are not directly driven by the host immune-system, e.g. the expression of surface sugars by bacteria. By sorting and sequencing IgA-coated and lectin-stained fractions from CD patients microbiota and healthy controls, we found that lectin-stained bacteria were distinct from IgA-coated bacteria, but still displayed specific differences between CD and healthy controls. To exploit the discriminatory potential of both, immunoglobulin coated bacteria and the altered surface sugar expression of bacteria in CD, we developed a multiplexed single cell-based analysis approach for intestinal microbiota. By multi-parameter microbiota flow cytometry (mMFC) we characterized the intestinal microbiota of 55 CD patients and 44 healthy controls for 11-parameters in total, comprising host-immunoglobulin coating and the presence of distinct surface sugar moieties. The data were analyzed by machine-learning to assess disease-specific marker patterns in the microbiota phenotype. mMFC captured detailed characteristics of CD microbiota and identified patterns to classify CD patients. In addition, we identified phenotypic signatures in the CD microbiota which not only reflected remission after 6 weeks of anti-TNF treatment, but were also able to predict remission before the start of an adalimumab treatment course in a pilot study. We here present the proof-of-concept demonstrating that multi-parameter single-cell bacterial phenotyping by mMFC could be a novel tool with high translational potential to expand current microbiome investigations by phenotyping of bacteria to identify disease- and therapy-associated cellular alterations and to reveal novel target properties of bacteria for functional assays and therapeutic approaches.
Keywords: Single-cell analysis; microbiota flow cytometry; microbiota phenotyping.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
