The fly route of extended-spectrum-β-lactamase-producing Enterobacteriaceae dissemination in a cattle farm: from the ecosystem to the molecular scale
- PMID: 39816254
- PMCID: PMC11732033
- DOI: 10.3389/frabi.2024.1367936
The fly route of extended-spectrum-β-lactamase-producing Enterobacteriaceae dissemination in a cattle farm: from the ecosystem to the molecular scale
Abstract
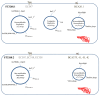
Introduction: This study aimed to understand the origin and to explain the maintenance of extended-spectrum β-lactamase (ESBL) Enterobacteriaceae isolated from food-producing animals in a third-generation cephalosporin (3GC)-free farm.
Methods: Culture and molecular approaches were used to test molecules other than 3GC such as antibiotics (tetracycline and oxytetracycline), antiparasitics (ivermectin, flumethrin, fenbendazol, and amitraz), heavy metal [arsenic, HNO3, aluminum, HNO3, cadmium (CdSO4), zinc (ZnCl2), copper (CuSO4), iron (FeCl3), and aluminum (Al2SO4)], and antioxidant (butylated hydroxytoluene) as sources of selective pressure. Whole-genome sequencing using short read (Illumina™) and long read (Nanopore™) technologies was performed on 34 genomes. In silico gene screening and comparative analyses were used to characterize the genetic determinants of resistance, their mobility, and the genomic relatedness among isolates.
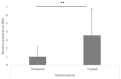
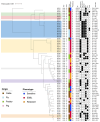
Results: Our analysis unveiled a low diversity among the animal ESBL-producing strains. Notably, E. coli ST3268 was recurrently isolated from both flies (n = 9) and cattle (n = 5). These E. coli ST3268/bla CTX-M-15/bla TEM-1B have accumulated multiple plasmids and genes, thereby representing a reservoir of resistance and virulence factors. Our findings suggest that flies could act as effective mechanical vectors for antimicrobial gene transfer and are capable of transporting resistant bacteria across different environments and to multiple hosts, facilitating the spread of pathogenic traits. A significantly higher mean minimum inhibitory concentration of oxytetracycline (841.4 ± 323.5 mg/L vs. 36.0 ± 52.6 mg/L, p = 0.0022) in ESBL E. coli than in non-ESBL E. coli and bla CTX-M-15 gene overexpression in oxytetracycline-treated vs. untreated ESBL E. coli (RQOxy = 3.593, p = 0.024) confirmed oxytetracycline as a source of selective pressure in ESBL E. coli.
Discussion: The occurrence of ESBL E. coli in a farm without 3GC use is probably due to an as yet undefined human origin of Enterobacteriaceae bla CTX-M-15 gene transmission to animals in close contact with cattle farm workers and the maintenance of the local ESBL E. coli reservoir by a high fly diversity and oxytetracycline selective pressure. These findings highlight the critical need for stringent vector control to mitigate antimicrobial resistance spread for preserving public health. Addressing this issue necessitates a multifaceted approach combining microbial genetics, vector ecology, and farm management practices.
Keywords: ESBL; Enterobacteriaceae; ST3268; oxytetracycline; selective pressure.
Copyright © 2024 Caderhoussin, Couvin, Gruel, Quétel, Pot, Arquet, Dereeper, Bambou, Talarmin and Ferdinand.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Collis R. M., Biggs P. J., Burgess S. A., Midwinter A. C., Brightwell G., Cookson A. L. (2022). Prevalence and distribution of extended-spectrum β-lactamase and AmpC-producing Escherichia coli in two New Zealand dairy farm environments. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.960748 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

