Regulatory T cell-associated gene signature correlates with prognostic risk and immune infiltration in patients with breast cancer
- PMID: 39816556
- PMCID: PMC11729763
- DOI: 10.21037/tcr-24-1118
Regulatory T cell-associated gene signature correlates with prognostic risk and immune infiltration in patients with breast cancer
Abstract
Background: Regulatory T cells (Tregs) play a pivotal role in the development, prognosis, and treatment of breast cancer. This study aimed to develop a Treg-associated gene signature that contributes to predict prognosis and therapy benefits in breast cancer.
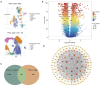
Methods: Treg-associated genes were screened based on single-cell RNA-sequencing (RNA-seq) in TISCH2 database and the bulk RNA-seq in The Cancer Genome Atlas (TCGA) database. Treg-associated gene signature was identified via survival analysis, univariate cox, least absolute shrinkage and selection operator (LASSO) and multivariable Cox regression analyses. Immune status was assessed using single-sample gene set enrichment analysis (ssGSEA) and Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) algorithms. Drug sensitivity was estimated using pRRophetic. Gene set enrichment analysis (GSEA) was conducted to explore the changed pathways.
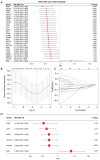
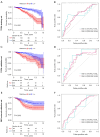
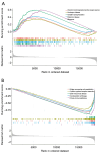
Results: A total of 169 genes were identified as Treg-associated genes, and close interactions existed among these genes. Kaplan-Meier (KM) survival and univariate cox revealed 29 prognostic genes (all P<0.05), and finally a six-gene prognostic signature including TBC1D4, PMAIP1, IFNG, LEF1, MZB1 and EZR was identified by LASSO and multivariable Cox. Based on this signature, patients in high-risk group exhibited a worse survival probability than those in low-risk group in the TCGA training dataset (P<0.001). Additionally, this signature showed a moderate predictive power for 1-, 3- and 5-year survival for breast cancer patients in both training dataset [area under the curve (AUC) =0.705, 0.678 and 0.668, respectively]. Similar predictive power for 1-, 3- and 5-year survival was also observed in validation datasets. Risk scores significantly differed between subgroups divided by clinicopathologic features, especially by molecular subtypes. Patients in high- and low-risk groups showed significant differences on infiltration abundance of multiple types of immune cells (such as, activated B cells/CD8+ T cells/CD4+ T cells), immune and stromal scores (all P<0.05). Moreover, sensitivity to 83 chemotherapeutic drugs such as lapatinib, methotrexate, and gefitinib were significantly differed between the two risk groups (all P<0.001).
Conclusions: This is the first to develop a Treg-associated gene signature for breast cancer, which could predict prognosis of patients and help to identify patients who might be benefit from immunotherapy and/or chemotherapy.
Keywords: Breast cancer; immune infiltration; prognostic signature; regulatory T cell (Treg).
2024 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1118/coif). The authors have no conflicts of interest to declare.
Figures



References
-
- Onkar SS, Carleton NM, Lucas PC, et al. The Great Immune Escape: Understanding the Divergent Immune Response in Breast Cancer Subtypes. Cancer Discov 2023;13:23-40. 10.1158/2159-8290.CD-22-0475 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
