Large-scale proteomic analysis of T. spiralis muscle-stage ESPs identifies a novel upstream motif for in silico prediction of secreted products
- PMID: 39816813
- PMCID: PMC11731790
- DOI: 10.3389/fpara.2023.1078443
Large-scale proteomic analysis of T. spiralis muscle-stage ESPs identifies a novel upstream motif for in silico prediction of secreted products
Abstract
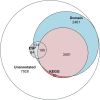
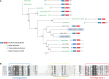
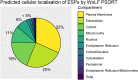
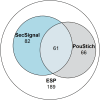
The Trichinella genus contains parasitic nematodes capable of infecting a wide range of hosts including mammals, birds and reptiles. Like other helminths, T. spiralis secretes a complex mixture of bioactive molecules capable of modulating its immediate surroundings and creating a hospitable environment for growth, survival and ultimately transmission. The constitution of these excretory-secretory products (ESPs) changes depending on the tissue niche and the specific stage of parasite development. Unique to T. spiralis is a true intracellular stage wherein larvae develop inside striated myotubes. Remarkably, the parasite larvae do not destroy the host cell but rather reprogram it to support their presence and growth. This transformation is largely mediated through stage-specific secretions released into the host cell cytoplasm. In this study, we apply state of the art proteomics and computational approaches to elucidate the composition and functions of muscle-stage T. spiralis ESPs. Moreover, we define a recurring, upstream motif associated with the stichosome, the main secretory organ of this worm, and can be used to predict secreted proteins across experimentally less tractable T. spiralis life cycle stages.
Keywords: ES; Trichinella; helminth; proteomics; secretions.
Copyright © 2023 Nash, Gregory, White, Protasio, Gygi, Selkirk, Weekes and Artavanis-Tsakonas.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

