Manipulation of mitochondrial poly(A) polymerase family proteins in Trypanosoma brucei impacts mRNA termini processing
- PMID: 39816830
- PMCID: PMC11732105
- DOI: 10.3389/fpara.2023.1298561
Manipulation of mitochondrial poly(A) polymerase family proteins in Trypanosoma brucei impacts mRNA termini processing
Abstract
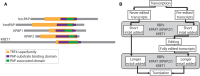
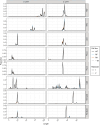
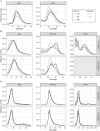
RNA-specific nucleotidyltransferases (rNTrs) add nontemplated nucleotides to the 3' end of RNA. Two noncanonical rNTRs that are thought to be poly(A) polymerases (PAPs) have been identified in the mitochondria of trypanosomes - KPAP1 and KPAP2. KPAP1 is the primary polymerase that adds adenines (As) to trypanosome mitochondrial mRNA 3' tails, while KPAP2 is a non-essential putative polymerase whose role in the mitochondria is ambiguous. Here, we elucidate the effects of manipulations of KPAP1 and KPAP2 on the 5' and 3' termini of transcripts and their 3' tails. Using glycerol gradients followed by immunoblotting, we present evidence that KPAP2 is found in protein complexes of up to about 1600 kDa. High-throughput sequencing of mRNA termini showed that KPAP2 overexpression subtly changes an edited transcript's 3' tails, though not in a way consistent with general PAP activity. Next, to identify possible roles of posttranslational modifications on KPAP1 regulation, we mutated two KPAP1 arginine methylation sites to either mimic methylation or hypomethylation. We assessed their effect on 3' mRNA tail characteristics and found that the two mutants generally had opposing effects, though some of these were transcript-specific. We present results suggesting that while methylation increases KPAP1 substrate binding and/or initial nucleotide additions, unmethylated KPAP1is more processive. We also present a comprehensive review of UTR termini, and evidence that tail addition activity may change as mRNA editing is initiated. Together, this work furthers our understanding of the role of KPAP1 and KPAP2 on trypanosome mitochondrial mRNA 3' tail addition, as well as provides more information on mRNA termini processing in general.
Keywords: African trypanosomiasis; Hidden Markov modelling; arginine methylation; mRNA tails; non-coding RNA; nucleotidyltransferases; site-directed mutagenesis.
Copyright © 2024 Smoniewski, Mirzavand Borujeni, Hampton, Petersen, Faacks, Salavati and Zimmer.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Targeted depletion of a mitochondrial nucleotidyltransferase suggests the presence of multiple enzymes that polymerize mRNA 3' tails in Trypanosoma brucei mitochondria.Mol Biochem Parasitol. 2007 Aug;154(2):158-69. doi: 10.1016/j.molbiopara.2007.04.014. Epub 2007 Apr 27. Mol Biochem Parasitol. 2007. PMID: 17543398 Free PMC article.
-
Additive and transcript-specific effects of KPAP1 and TbRND activities on 3' non-encoded tail characteristics and mRNA stability in Trypanosoma brucei.PLoS One. 2012;7(5):e37639. doi: 10.1371/journal.pone.0037639. Epub 2012 May 21. PLoS One. 2012. PMID: 22629436 Free PMC article.
-
3' adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria.EMBO J. 2008 Jun 4;27(11):1596-608. doi: 10.1038/emboj.2008.87. Epub 2008 May 8. EMBO J. 2008. PMID: 18464794 Free PMC article.
-
Mitochondrial RNA quality control in trypanosomes.Wiley Interdiscip Rev RNA. 2021 May;12(3):e1638. doi: 10.1002/wrna.1638. Epub 2020 Dec 16. Wiley Interdiscip Rev RNA. 2021. PMID: 33331073 Free PMC article. Review.
-
Dynamic RNA holo-editosomes with subcomplex variants: Insights into the control of trypanosome editing.Wiley Interdiscip Rev RNA. 2018 Nov;9(6):e1502. doi: 10.1002/wrna.1502. Epub 2018 Aug 12. Wiley Interdiscip Rev RNA. 2018. PMID: 30101566 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

