Structure-Based Optimization of Pyridone α-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease
- PMID: 39817813
- PMCID: PMC11831675
- DOI: 10.1021/acs.jmedchem.4c02172
Structure-Based Optimization of Pyridone α-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease
Abstract
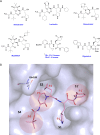
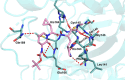
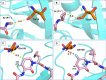
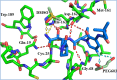
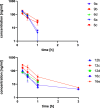
The main protease Mpro is a clinically validated target to treat infections by the coronavirus SARS-CoV-2. Among the first reported Mpro inhibitors was the peptidomimetic α-ketoamide 13b, whose cocrystal structure with Mpro paved the way for multiple lead-finding studies. We established structure-activity relationships for the 13b series by modifying residues at the P1', P3, and P4 sites. Guided by cocrystal structures, we reduced the P1' substituent size to better fill the pocket and added a fluorine substituent to the pyridone ring, enabling a new hydrogen bond with Gln189 in P3. Among 22 novel analogues, 6d and 12d inhibited Mpro with IC50s of 110 nM and 40 nM, improving the potency of 13b by up to 9.5-fold. Compound 6d had pronounced antiviral activity with an EC50 of 1.6 μM and was stable in plasma and microsomes. The study illustrates the potential of structure-based design to systematically improve peptidomimetic α-ketoamides.
Conflict of interest statement
The authors declare the following competing financial interest(s): The University of Lbeck has been granted a patent (DE 10 2020 103 516.0) covering the pyridone-containing inhibitors.
Figures
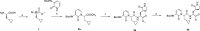
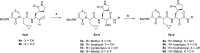
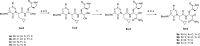
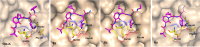
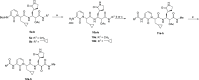
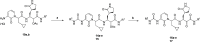
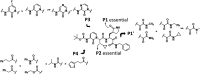
Similar articles
-
Site mapping and small molecule blind docking reveal a possible target site on the SARS-CoV-2 main protease dimer interface.Comput Biol Chem. 2020 Dec;89:107372. doi: 10.1016/j.compbiolchem.2020.107372. Epub 2020 Sep 5. Comput Biol Chem. 2020. PMID: 32911432 Free PMC article.
-
Exploring covalent inhibitors of SARS-CoV-2 main protease: from peptidomimetics to novel scaffolds.J Enzyme Inhib Med Chem. 2025 Dec;40(1):2460045. doi: 10.1080/14756366.2025.2460045. Epub 2025 Feb 6. J Enzyme Inhib Med Chem. 2025. PMID: 39912405 Free PMC article. Review.
-
Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors.Science. 2020 Apr 24;368(6489):409-412. doi: 10.1126/science.abb3405. Epub 2020 Mar 20. Science. 2020. PMID: 32198291 Free PMC article.
-
Synthesis and biological investigation of peptidomimetic SARS-CoV-2 main protease inhibitors bearing quinoline-based heterocycles at P3.Arch Pharm (Weinheim). 2025 Jan;358(1):e2400812. doi: 10.1002/ardp.202400812. Arch Pharm (Weinheim). 2025. PMID: 39873316
-
Design of inhibitors of SARS-CoV-2 papain-like protease deriving from GRL0617: Structure-activity relationships.Bioorg Med Chem. 2024 Nov 1;113:117909. doi: 10.1016/j.bmc.2024.117909. Epub 2024 Sep 11. Bioorg Med Chem. 2024. PMID: 39288705 Review.
References
-
- WHO , 2024. https://data.who.int/dashboards/covid19/cases.n=o (accessed Aug 30, 2024).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

